| 1 |
|
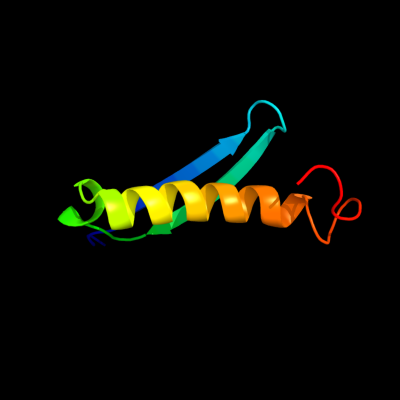
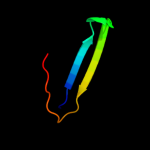
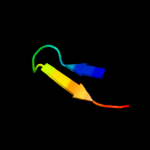
PDB 1nei chain A
Region: 1 - 60
Aligned: 60
Modelled: 60
Confidence: 100.0%
Identity: 100%
Fold: Hypothetical protein YoaG
Superfamily: Hypothetical protein YoaG
Family: Hypothetical protein YoaG
Phyre2
| 2 |
|
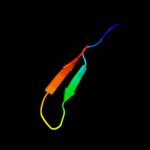
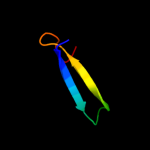
PDB 1e2w chain A domain 2
Region: 3 - 33
Aligned: 30
Modelled: 31
Confidence: 36.2%
Identity: 27%
Fold: Barrel-sandwich hybrid
Superfamily: Rudiment single hybrid motif
Family: Cytochrome f, small domain
Phyre2
| 3 |
|
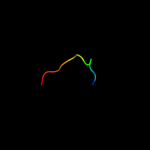
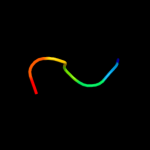
PDB 1czp chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 20.8%
Identity: 29%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin-related
Phyre2
| 4 |
|
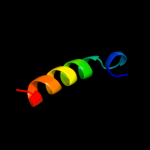
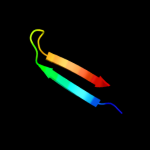
PDB 1ci3 chain M domain 2
Region: 3 - 33
Aligned: 30
Modelled: 31
Confidence: 16.1%
Identity: 17%
Fold: Barrel-sandwich hybrid
Superfamily: Rudiment single hybrid motif
Family: Cytochrome f, small domain
Phyre2
| 5 |
|
PDB 2ww1 chain B
Region: 26 - 47
Aligned: 22
Modelled: 22
Confidence: 15.7%
Identity: 32%
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt39902 from bacteroides thetaiotaomicron vpi-5482 in complex with3 thiomannobioside
Phyre2
| 6 |
|
PDB 1hcz chain A domain 2
Region: 3 - 33
Aligned: 31
Modelled: 31
Confidence: 12.9%
Identity: 32%
Fold: Barrel-sandwich hybrid
Superfamily: Rudiment single hybrid motif
Family: Cytochrome f, small domain
Phyre2
| 7 |
|
PDB 1pfd chain A
Region: 4 - 20
Aligned: 17
Modelled: 17
Confidence: 11.3%
Identity: 29%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin-related
Phyre2
| 8 |
|
PDB 1off chain A
Region: 4 - 20
Aligned: 17
Modelled: 17
Confidence: 11.2%
Identity: 29%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin-related
Phyre2
| 9 |
|
PDB 1wcu chain A
Region: 46 - 57
Aligned: 12
Modelled: 12
Confidence: 10.3%
Identity: 42%
PDB header:carbohydrate binding
Chain: A: PDB Molecule:non-catalytic protein 1;
PDBTitle: cbm29_1, a family 29 carbohydrate binding module from2 piromyces equi
Phyre2
| 10 |
|
PDB 1j8y chain F domain 1
Region: 24 - 47
Aligned: 24
Modelled: 24
Confidence: 10.0%
Identity: 21%
Fold: Four-helical up-and-down bundle
Superfamily: Domain of the SRP/SRP receptor G-proteins
Family: Domain of the SRP/SRP receptor G-proteins
Phyre2
| 11 |
|
PDB 1rj9 chain B domain 1
Region: 24 - 47
Aligned: 24
Modelled: 24
Confidence: 9.5%
Identity: 38%
Fold: Four-helical up-and-down bundle
Superfamily: Domain of the SRP/SRP receptor G-proteins
Family: Domain of the SRP/SRP receptor G-proteins
Phyre2
| 12 |
|
PDB 1gaq chain B
Region: 4 - 20
Aligned: 17
Modelled: 17
Confidence: 9.3%
Identity: 29%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin-related
Phyre2
| 13 |
|
PDB 2wvy chain A
Region: 26 - 47
Aligned: 22
Modelled: 22
Confidence: 9.0%
Identity: 36%
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-1,2-mannosidase;
PDBTitle: structure of the family gh92 inverting mannosidase bt21992 from bacteroides thetaiotaomicron vpi-5482
Phyre2
| 14 |
|
PDB 1ls1 chain A domain 1
Region: 24 - 47
Aligned: 24
Modelled: 24
Confidence: 8.7%
Identity: 38%
Fold: Four-helical up-and-down bundle
Superfamily: Domain of the SRP/SRP receptor G-proteins
Family: Domain of the SRP/SRP receptor G-proteins
Phyre2
| 15 |
|
PDB 1gwm chain A
Region: 46 - 57
Aligned: 12
Modelled: 12
Confidence: 8.6%
Identity: 17%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 29 carbohydrate binding module, CBM29
Phyre2
| 16 |
|
PDB 1wgw chain A
Region: 24 - 47
Aligned: 24
Modelled: 24
Confidence: 8.6%
Identity: 29%
Fold: Four-helical up-and-down bundle
Superfamily: Domain of the SRP/SRP receptor G-proteins
Family: Domain of the SRP/SRP receptor G-proteins
Phyre2
| 17 |
|
PDB 4fxc chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 8.6%
Identity: 29%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: 2Fe-2S ferredoxin-like
Family: 2Fe-2S ferredoxin-related
Phyre2
| 18 |
|
PDB 2wuk chain D
Region: 47 - 53
Aligned: 7
Modelled: 7
Confidence: 7.2%
Identity: 43%
PDB header:cell cycle
Chain: D: PDB Molecule:septum site-determining protein diviva;
PDBTitle: diviva n-terminal domain, f17a mutant
Phyre2
| 19 |
|
PDB 3ku7 chain B
Region: 37 - 49
Aligned: 13
Modelled: 13
Confidence: 7.0%
Identity: 23%
PDB header:cell cycle
Chain: B: PDB Molecule:cell division topological specificity factor;
PDBTitle: crystal structure of helicobacter pylori mine, a cell division2 topological specificity factor
Phyre2
| 20 |
|
PDB 3emi chain A
Region: 5 - 22
Aligned: 18
Modelled: 18
Confidence: 6.8%
Identity: 44%
PDB header:cell adhesion
Chain: A: PDB Molecule:hia (adhesin);
PDBTitle: crystal structure of hia 307-422 non-adhesive domain
Phyre2
| 21 |
|
| 22 |
|