| 1 |
|
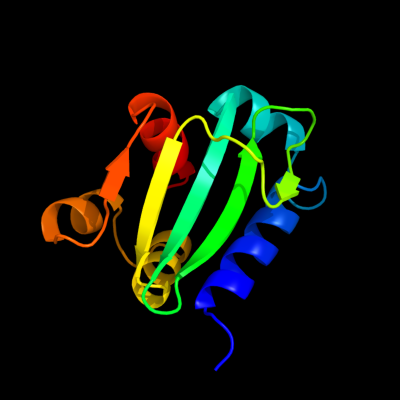
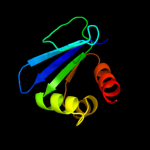
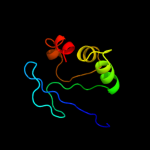
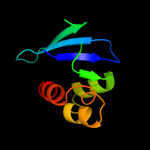
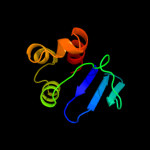
PDB 1ikt chain A
Region: 28 - 135
Aligned: 106
Modelled: 108
Confidence: 99.5%
Identity: 17%
Fold: SCP-like
Superfamily: SCP-like
Family: Sterol carrier protein, SCP
Phyre2
| 2 |
|
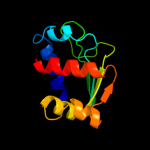
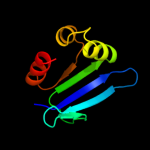
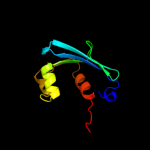
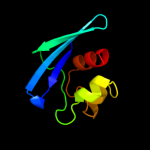
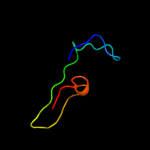
PDB 1c44 chain A
Region: 25 - 133
Aligned: 106
Modelled: 109
Confidence: 99.5%
Identity: 26%
Fold: SCP-like
Superfamily: SCP-like
Family: Sterol carrier protein, SCP
Phyre2
| 3 |
|
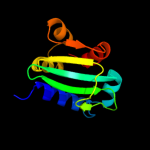
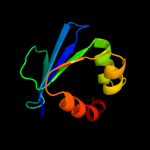
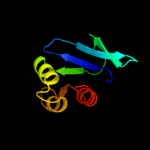
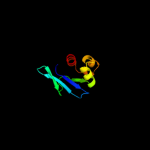
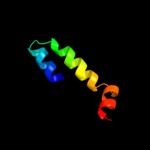
PDB 3bn8 chain A
Region: 57 - 135
Aligned: 78
Modelled: 79
Confidence: 99.4%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:putative sterol carrier protein 2;
PDBTitle: crystal structure of a putative sterol carrier protein type 2 (af1534)2 from archaeoglobus fulgidus dsm 4304 at 2.11 a resolution
Phyre2
| 4 |
|
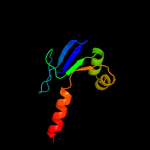
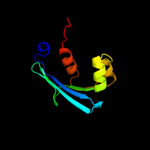
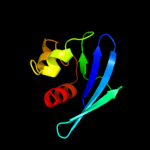
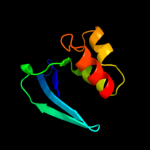
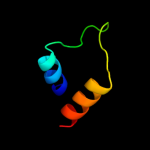
PDB 3bkr chain A
Region: 55 - 134
Aligned: 80
Modelled: 80
Confidence: 99.4%
Identity: 19%
PDB header:lipid binding protein
Chain: A: PDB Molecule:sterol carrier protein-2 like-3;
PDBTitle: crystal structure of sterol carrier protein-2 like-3 (scp2-2 l3) from aedes aegypti
Phyre2
| 5 |
|
PDB 3bdq chain B
Region: 55 - 132
Aligned: 78
Modelled: 78
Confidence: 99.4%
Identity: 18%
PDB header:lipid transport
Chain: B: PDB Molecule:sterol carrier protein 2-like 2;
PDBTitle: room tempreture crystal structure of sterol carrier protein-2 2 like-2
Phyre2
| 6 |
|
PDB 1pz4 chain A
Region: 55 - 135
Aligned: 80
Modelled: 81
Confidence: 99.3%
Identity: 16%
Fold: SCP-like
Superfamily: SCP-like
Family: Sterol carrier protein, SCP
Phyre2
| 7 |
|
PDB 2cfu chain A domain 1
Region: 47 - 138
Aligned: 92
Modelled: 92
Confidence: 99.2%
Identity: 16%
Fold: SCP-like
Superfamily: SCP-like
Family: Alkylsulfatase C-terminal domain-like
Phyre2
| 8 |
|
PDB 1wfr chain A
Region: 63 - 135
Aligned: 73
Modelled: 73
Confidence: 99.0%
Identity: 14%
Fold: SCP-like
Superfamily: SCP-like
Family: Sterol carrier protein, SCP
Phyre2
| 9 |
|
PDB 2cfu chain A
Region: 46 - 138
Aligned: 93
Modelled: 93
Confidence: 98.2%
Identity: 16%
PDB header:hydrolase
Chain: A: PDB Molecule:sdsa1;
PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
Phyre2
| 10 |
|
PDB 2i00 chain A domain 1
Region: 54 - 132
Aligned: 77
Modelled: 79
Confidence: 96.6%
Identity: 14%
Fold: SCP-like
Superfamily: SCP-like
Family: EF1021 C-terminal domain-like
Phyre2
| 11 |
|
PDB 2ozg chain A domain 1
Region: 57 - 132
Aligned: 72
Modelled: 76
Confidence: 96.2%
Identity: 22%
Fold: SCP-like
Superfamily: SCP-like
Family: EF1021 C-terminal domain-like
Phyre2
| 12 |
|
PDB 2hv2 chain A domain 1
Region: 57 - 132
Aligned: 74
Modelled: 76
Confidence: 95.5%
Identity: 18%
Fold: SCP-like
Superfamily: SCP-like
Family: EF1021 C-terminal domain-like
Phyre2
| 13 |
|
PDB 2ozg chain A
Region: 57 - 132
Aligned: 72
Modelled: 76
Confidence: 95.2%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:gcn5-related n-acetyltransferase;
PDBTitle: crystal structure of gcn5-related n-acetyltransferase (yp_325469.1)2 from anabaena variabilis atcc 29413 at 2.00 a resolution
Phyre2
| 14 |
|
PDB 2i00 chain D
Region: 54 - 131
Aligned: 76
Modelled: 78
Confidence: 92.9%
Identity: 14%
PDB header:transferase
Chain: D: PDB Molecule:acetyltransferase, gnat family;
PDBTitle: crystal structure of acetyltransferase (gnat family) from enterococcus2 faecalis
Phyre2
| 15 |
|
PDB 2hv2 chain D
Region: 57 - 132
Aligned: 72
Modelled: 76
Confidence: 92.5%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of conserved protein of unknown function from2 enterococcus faecalis v583 at 2.4 a resolution, probable n-3 acyltransferase
Phyre2
| 16 |
|
PDB 3r1k chain A
Region: 57 - 132
Aligned: 72
Modelled: 76
Confidence: 79.7%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:enhanced intracellular survival protein;
PDBTitle: crystal structure of acetyltransferase eis from mycobacterium2 tuberculosis h37rv in complex with coa and an acetamide moiety
Phyre2
| 17 |
|
PDB 3n7z chain D
Region: 58 - 132
Aligned: 70
Modelled: 75
Confidence: 78.2%
Identity: 14%
PDB header:transferase
Chain: D: PDB Molecule:acetyltransferase, gnat family;
PDBTitle: crystal structure of acetyltransferase from bacillus anthracis
Phyre2
| 18 |
|
PDB 3d33 chain B
Region: 53 - 91
Aligned: 32
Modelled: 37
Confidence: 9.2%
Identity: 25%
PDB header:unknown function
Chain: B: PDB Molecule:domain of unknown function with an immunoglobulin-like
PDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
Phyre2
| 19 |
|
PDB 1y76 chain A domain 1
Region: 2 - 39
Aligned: 38
Modelled: 38
Confidence: 8.5%
Identity: 24%
Fold: L27 domain
Superfamily: L27 domain
Family: L27 domain
Phyre2
| 20 |
|
PDB 1xn8 chain A
Region: 118 - 154
Aligned: 37
Modelled: 37
Confidence: 7.5%
Identity: 27%
Fold: Hypothetical protein YqbG
Superfamily: Hypothetical protein YqbG
Family: Hypothetical protein YqbG
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|