| 1 |
|
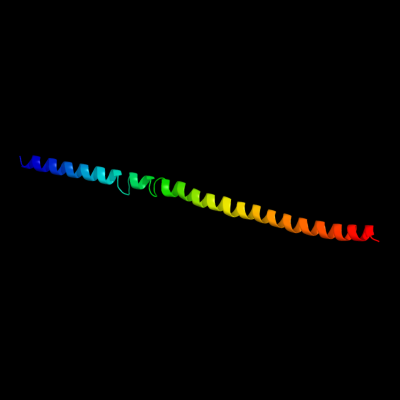
PDB 3u59 chain C
Region: 20 - 105
Aligned: 82
Modelled: 86
Confidence: 68.4%
Identity: 12%
PDB header:contractile protein
Chain: C: PDB Molecule:tropomyosin beta chain;
PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
Phyre2
| 2 |
|
PDB 2gl2 chain B
Region: 16 - 104
Aligned: 85
Modelled: 89
Confidence: 45.2%
Identity: 12%
PDB header:cell adhesion
Chain: B: PDB Molecule:adhesion a;
PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
Phyre2
| 3 |
|
PDB 4a55 chain B
Region: 44 - 105
Aligned: 60
Modelled: 62
Confidence: 34.6%
Identity: 12%
PDB header:transferase
Chain: B: PDB Molecule:phosphatidylinositol 3-kinase regulatory subunit alpha;
PDBTitle: crystal structure of p110alpha in complex with ish2 of p85alpha and2 the inhibitor pik-108
Phyre2
| 4 |
|
PDB 2ch7 chain A
Region: 11 - 109
Aligned: 97
Modelled: 99
Confidence: 29.1%
Identity: 6%
PDB header:chemotaxis
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
Phyre2
| 5 |
|
PDB 1vp7 chain A
Region: 59 - 104
Aligned: 46
Modelled: 46
Confidence: 24.0%
Identity: 7%
Fold: Spectrin repeat-like
Superfamily: XseB-like
Family: XseB-like
Phyre2
| 6 |
|
PDB 2e7s chain M
Region: 46 - 105
Aligned: 58
Modelled: 60
Confidence: 16.0%
Identity: 12%
PDB header:endocytosis/exocytosis
Chain: M: PDB Molecule:rab guanine nucleotide exchange factor sec2;
PDBTitle: crystal structure of the yeast sec2p gef domain
Phyre2
| 7 |
|
PDB 3hnw chain B
Region: 13 - 105
Aligned: 81
Modelled: 93
Confidence: 15.0%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
Phyre2
| 8 |
|
PDB 1vp7 chain B
Region: 59 - 104
Aligned: 46
Modelled: 46
Confidence: 11.7%
Identity: 7%
Fold: Spectrin repeat-like
Superfamily: XseB-like
Family: XseB-like
Phyre2
| 9 |
|
PDB 1ujw chain B
Region: 58 - 103
Aligned: 46
Modelled: 46
Confidence: 8.4%
Identity: 13%
PDB header:transport protein/hydrolase
Chain: B: PDB Molecule:colicin e3;
PDBTitle: structure of the complex between btub and colicin e3 receptor binding2 domain
Phyre2
| 10 |
|
PDB 2nps chain C
Region: 22 - 84
Aligned: 59
Modelled: 63
Confidence: 7.7%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:vesicle transport through interaction with t-
PDBTitle: crystal structure of the early endosomal snare complex
Phyre2
| 11 |
|
PDB 3ajw chain A
Region: 58 - 109
Aligned: 52
Modelled: 52
Confidence: 7.7%
Identity: 17%
PDB header:protein transport
Chain: A: PDB Molecule:flagellar flij protein;
PDBTitle: structure of flij, a soluble component of flagellar type iii export2 apparatus
Phyre2
| 12 |
|
PDB 2voy chain K
Region: 1 - 11
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 36%
PDB header:hydrolase
Chain: K: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 13 |
|
PDB 3ol1 chain A
Region: 23 - 104
Aligned: 78
Modelled: 82
Confidence: 6.3%
Identity: 19%
PDB header:structural protein
Chain: A: PDB Molecule:vimentin;
PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
Phyre2
| 14 |
|
PDB 1gl2 chain C
Region: 22 - 83
Aligned: 58
Modelled: 62
Confidence: 6.0%
Identity: 10%
PDB header:membrane protein
Chain: C: PDB Molecule:vesicle transport v-snare protein vti1-like 1;
PDBTitle: crystal structure of an endosomal snare core complex
Phyre2