| 1 |
|
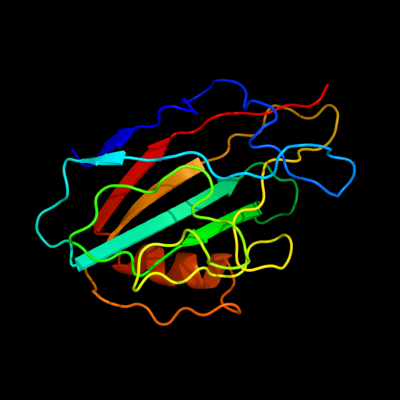
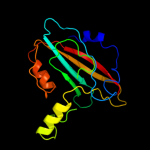
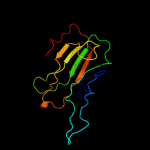
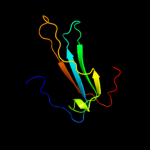
PDB 1fux chain A
Region: 21 - 183
Aligned: 163
Modelled: 163
Confidence: 100.0%
Identity: 99%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Prokaryotic PEBP-like proteins
Phyre2
| 2 |
|
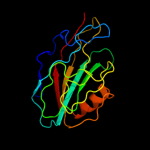
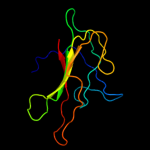
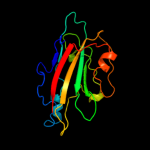
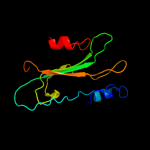
PDB 1fjj chain A
Region: 21 - 180
Aligned: 158
Modelled: 158
Confidence: 100.0%
Identity: 51%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Prokaryotic PEBP-like proteins
Phyre2
| 3 |
|
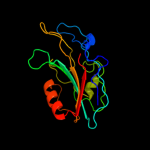
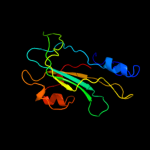
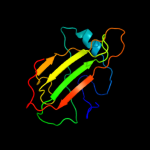
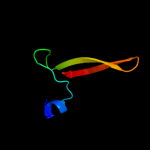
PDB 3n08 chain A
Region: 21 - 182
Aligned: 146
Modelled: 157
Confidence: 100.0%
Identity: 36%
PDB header:phosphatidylethanolamine-binding protein
Chain: A: PDB Molecule:putative phosphatidylethanolamine-binding protein (pebp);
PDBTitle: crystal structure of a putative phosphatidylethanolamine-binding2 protein (pebp) homolog ct736 from chlamydia trachomatis d/uw-3/cx
Phyre2
| 4 |
|
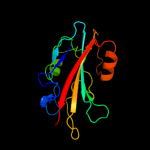
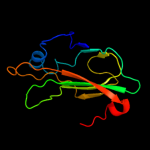
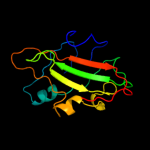
PDB 2evv chain D
Region: 21 - 182
Aligned: 153
Modelled: 157
Confidence: 100.0%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein hp0218;
PDBTitle: crystal structure of the pebp-like protein of unknown function hp02182 from helicobacter pylori
Phyre2
| 5 |
|
PDB 2r77 chain A
Region: 26 - 145
Aligned: 104
Modelled: 104
Confidence: 99.8%
Identity: 24%
PDB header:lipid binding protein
Chain: A: PDB Molecule:phosphatidylethanolamine-binding protein, putative;
PDBTitle: crystal structure of phosphatidylethanolamine-binding protein,2 pfl0955c, from plasmodium falciparum
Phyre2
| 6 |
|
PDB 1wpx chain B domain 1
Region: 29 - 180
Aligned: 128
Modelled: 140
Confidence: 99.8%
Identity: 22%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Phosphatidylethanolamine binding protein
Phyre2
| 7 |
|
PDB 2gzq chain A
Region: 21 - 156
Aligned: 112
Modelled: 118
Confidence: 99.8%
Identity: 23%
PDB header:lipid binding protein
Chain: A: PDB Molecule:phosphatidylethanolamine-binding protein;
PDBTitle: phosphatidylethanolamine-binding protein from plasmodium vivax
Phyre2
| 8 |
|
PDB 2jyz chain A
Region: 12 - 158
Aligned: 118
Modelled: 128
Confidence: 99.8%
Identity: 30%
PDB header:unknown function
Chain: A: PDB Molecule:cg7054-pa;
PDBTitle: cg7054 solution structure
Phyre2
| 9 |
|
PDB 1wkp chain A
Region: 17 - 183
Aligned: 134
Modelled: 154
Confidence: 99.7%
Identity: 22%
PDB header:signaling protein
Chain: A: PDB Molecule:flowering locus t protein;
PDBTitle: flowering locus t (ft) from arabidopsis thaliana
Phyre2
| 10 |
|
PDB 1kn3 chain A
Region: 16 - 155
Aligned: 109
Modelled: 119
Confidence: 99.7%
Identity: 28%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Phosphatidylethanolamine binding protein
Phyre2
| 11 |
|
PDB 2qyq chain A domain 1
Region: 13 - 155
Aligned: 112
Modelled: 129
Confidence: 99.7%
Identity: 29%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Phosphatidylethanolamine binding protein
Phyre2
| 12 |
|
PDB 1a44 chain A
Region: 53 - 155
Aligned: 81
Modelled: 82
Confidence: 99.7%
Identity: 26%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Phosphatidylethanolamine binding protein
Phyre2
| 13 |
|
PDB 1qou chain A
Region: 29 - 165
Aligned: 105
Modelled: 115
Confidence: 99.7%
Identity: 20%
Fold: PEBP-like
Superfamily: PEBP-like
Family: Phosphatidylethanolamine binding protein
Phyre2
| 14 |
|
PDB 3ks7 chain D
Region: 46 - 87
Aligned: 42
Modelled: 42
Confidence: 45.6%
Identity: 26%
PDB header:hydrolase
Chain: D: PDB Molecule:putative putative pngase f;
PDBTitle: crystal structure of putative peptide:n-glycosidase f (pngase f)2 (yp_210507.1) from bacteroides fragilis nctc 9343 at 2.30 a3 resolution
Phyre2
| 15 |
|
PDB 1qcs chain A domain 1
Region: 103 - 143
Aligned: 38
Modelled: 41
Confidence: 11.1%
Identity: 21%
Fold: Double psi beta-barrel
Superfamily: ADC-like
Family: Cdc48 N-terminal domain-like
Phyre2
| 16 |
|
PDB 1ljz chain B
Region: 80 - 97
Aligned: 18
Modelled: 18
Confidence: 10.7%
Identity: 33%
PDB header:receptor, toxin
Chain: B: PDB Molecule:acetylcholine receptor protein;
PDBTitle: nmr structure of an achr-peptide (torpedo californica,2 alpha-subunit residues 182-202) in complex with alpha-3 bungarotoxin
Phyre2
| 17 |
|
PDB 3pnr chain B
Region: 69 - 90
Aligned: 21
Modelled: 22
Confidence: 9.9%
Identity: 24%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:pbicp-c;
PDBTitle: structure of pbicp-c in complex with falcipain-2
Phyre2
| 18 |
|
PDB 3kml chain B
Region: 55 - 70
Aligned: 16
Modelled: 16
Confidence: 7.2%
Identity: 38%
PDB header:viral protein
Chain: B: PDB Molecule:coat protein;
PDBTitle: circular permutant of the tobacco mosaic virus
Phyre2