| 1 |
|
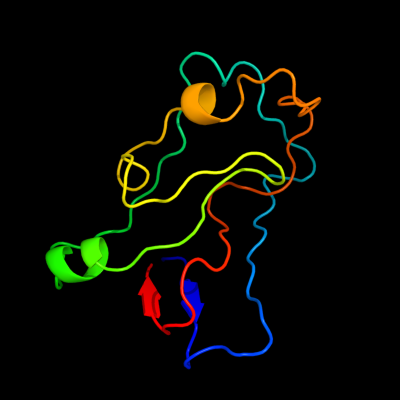
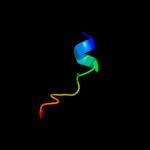
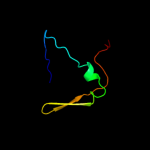
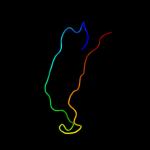
PDB 1wmx chain A
Region: 42 - 152
Aligned: 103
Modelled: 111
Confidence: 19.1%
Identity: 13%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 30 carbohydrate binding module, CBM30 (PKD repeat)
Phyre2
| 2 |
|
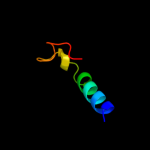
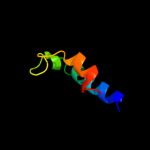
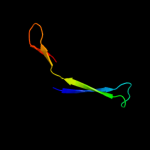
PDB 3alx chain B
Region: 147 - 211
Aligned: 44
Modelled: 45
Confidence: 16.9%
Identity: 16%
PDB header:viral protein/membrane protein
Chain: B: PDB Molecule:hemagglutinin, cdw150;
PDBTitle: crystal structure of the measles virus hemagglutinin bound to its2 cellular receptor slam (mv-h(l482r)-slam(n102h/r108y) fusion)
Phyre2
| 3 |
|
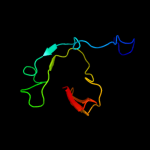
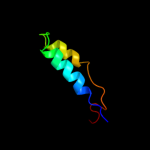
PDB 1jmx chain A domain 2
Region: 109 - 139
Aligned: 31
Modelled: 31
Confidence: 15.5%
Identity: 16%
Fold: Cytochrome c
Superfamily: Cytochrome c
Family: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
Phyre2
| 4 |
|
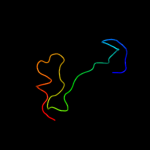
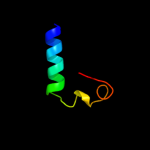
PDB 2kxo chain A
Region: 3 - 17
Aligned: 15
Modelled: 15
Confidence: 15.3%
Identity: 33%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division topological specificity factor;
PDBTitle: solution nmr structure of the cell division regulator mine protein2 from neisseria gonorrhoeae
Phyre2
| 5 |
|
PDB 1pby chain A domain 2
Region: 109 - 139
Aligned: 31
Modelled: 31
Confidence: 12.3%
Identity: 19%
Fold: Cytochrome c
Superfamily: Cytochrome c
Family: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
Phyre2
| 6 |
|
PDB 1gla chain F
Region: 3 - 83
Aligned: 80
Modelled: 81
Confidence: 9.9%
Identity: 18%
Fold: Barrel-sandwich hybrid
Superfamily: Duplicated hybrid motif
Family: Glucose permease-like
Phyre2
| 7 |
|
PDB 2dhz chain A
Region: 170 - 209
Aligned: 40
Modelled: 40
Confidence: 8.6%
Identity: 20%
PDB header:signaling protein
Chain: A: PDB Molecule:rap guanine nucleotide exchange factor (gef)-
PDBTitle: solution structure of the ra domain in the human link2 guanine nucleotide exchange factor ii (link-gefii)
Phyre2
| 8 |
|
PDB 3pmg chain A domain 4
Region: 98 - 149
Aligned: 51
Modelled: 52
Confidence: 7.7%
Identity: 10%
Fold: TBP-like
Superfamily: Phosphoglucomutase, C-terminal domain
Family: Phosphoglucomutase, C-terminal domain
Phyre2
| 9 |
|
PDB 1pby chain A
Region: 109 - 148
Aligned: 40
Modelled: 40
Confidence: 6.3%
Identity: 20%
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinohemoprotein amine dehydrogenase 60 kda
PDBTitle: structure of the phenylhydrazine adduct of the2 quinohemoprotein amine dehydrogenase from paracoccus3 denitrificans at 1.7 a resolution
Phyre2
| 10 |
|
PDB 2bid chain A
Region: 87 - 131
Aligned: 45
Modelled: 45
Confidence: 5.8%
Identity: 9%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 11 |
|
PDB 1jmx chain A
Region: 109 - 139
Aligned: 31
Modelled: 31
Confidence: 5.5%
Identity: 16%
PDB header:oxidoreductase
Chain: A: PDB Molecule:amine dehydrogenase;
PDBTitle: crystal structure of a quinohemoprotein amine dehydrogenase2 from pseudomonas putida
Phyre2
| 12 |
|
PDB 2ewc chain A domain 1
Region: 5 - 38
Aligned: 33
Modelled: 34
Confidence: 5.3%
Identity: 21%
Fold: Bacillus chorismate mutase-like
Superfamily: YjgF-like
Family: YjgF/L-PSP
Phyre2
| 13 |
|
PDB 2jeu chain A
Region: 131 - 167
Aligned: 37
Modelled: 37
Confidence: 5.2%
Identity: 8%
PDB header:transcription
Chain: A: PDB Molecule:regulatory protein e2;
PDBTitle: transcription activator structure reveals redox control of2 a replication initiation reaction
Phyre2