| 1 |
|
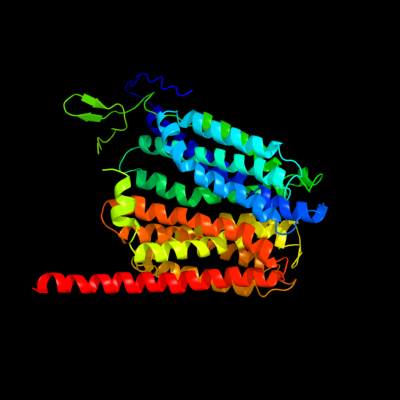
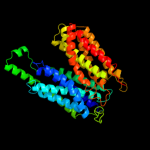
PDB 1pw4 chain A
Region: 28 - 468
Aligned: 426
Modelled: 431
Confidence: 100.0%
Identity: 18%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
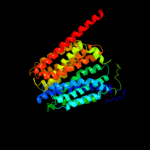
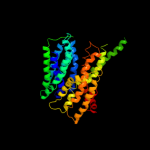
PDB 1pv7 chain A
Region: 41 - 469
Aligned: 416
Modelled: 429
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 3 |
|
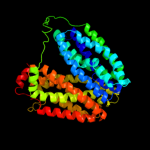
PDB 2gfp chain A
Region: 51 - 445
Aligned: 370
Modelled: 377
Confidence: 100.0%
Identity: 16%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
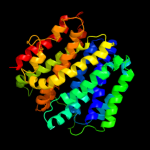
PDB 3o7p chain A
Region: 45 - 455
Aligned: 394
Modelled: 406
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 53 - 451
Aligned: 399
Modelled: 399
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 418 - 471
Aligned: 54
Modelled: 54
Confidence: 34.6%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 3b60 chain A domain 2
Region: 35 - 115
Aligned: 81
Modelled: 81
Confidence: 22.7%
Identity: 9%
Fold: ABC transporter transmembrane region
Superfamily: ABC transporter transmembrane region
Family: ABC transporter transmembrane region
Phyre2
| 8 |
|
PDB 2pkg chain C domain 1
Region: 8 - 25
Aligned: 18
Modelled: 18
Confidence: 18.5%
Identity: 33%
Fold: T-antigen specific domain-like
Superfamily: T-antigen specific domain-like
Family: T-antigen specific domain-like
Phyre2
| 9 |
|
PDB 3hd6 chain A
Region: 364 - 471
Aligned: 108
Modelled: 108
Confidence: 14.8%
Identity: 15%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 10 |
|
PDB 1pf4 chain A domain 2
Region: 35 - 115
Aligned: 81
Modelled: 81
Confidence: 10.7%
Identity: 9%
Fold: ABC transporter transmembrane region
Superfamily: ABC transporter transmembrane region
Family: ABC transporter transmembrane region
Phyre2
| 11 |
|
PDB 3b9y chain A
Region: 364 - 471
Aligned: 108
Modelled: 108
Confidence: 10.4%
Identity: 17%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 12 |
|
PDB 2knc chain A
Region: 430 - 467
Aligned: 38
Modelled: 38
Confidence: 10.1%
Identity: 18%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 13 |
|
PDB 2g9p chain A
Region: 104 - 117
Aligned: 14
Modelled: 14
Confidence: 9.6%
Identity: 21%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 14 |
|
PDB 3pro chain C domain 1
Region: 72 - 113
Aligned: 42
Modelled: 42
Confidence: 9.4%
Identity: 12%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 15 |
|
PDB 1fc3 chain A
Region: 42 - 87
Aligned: 45
Modelled: 46
Confidence: 7.6%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: Spo0A
Phyre2
| 16 |
|
PDB 1lq1 chain A
Region: 42 - 87
Aligned: 45
Modelled: 46
Confidence: 7.1%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: Spo0A
Phyre2
| 17 |
|
PDB 2aj2 chain A
Region: 3 - 18
Aligned: 16
Modelled: 16
Confidence: 6.5%
Identity: 25%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical upf0301 protein vc0467;
PDBTitle: x-ray crystal structure of protein vc0467 from vibrio2 cholerae. northeast structural genomics consortium target3 vcr8.
Phyre2
| 18 |
|
PDB 3arc chain T
Region: 435 - 459
Aligned: 25
Modelled: 25
Confidence: 6.0%
Identity: 12%
PDB header:electron transport, photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 19 |
|
PDB 3a0b chain T
Region: 435 - 459
Aligned: 25
Modelled: 25
Confidence: 6.0%
Identity: 12%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 20 |
|
PDB 2oar chain A domain 1
Region: 402 - 471
Aligned: 70
Modelled: 70
Confidence: 5.6%
Identity: 16%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 21 |
|
| 22 |
|