| 1 |
|
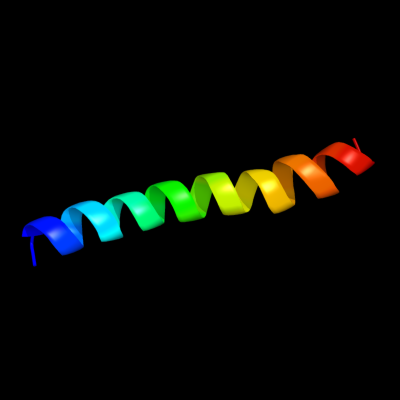
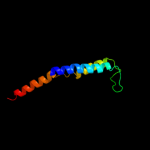
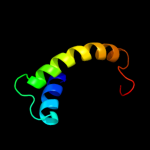
PDB 2knc chain A
Region: 332 - 363
Aligned: 31
Modelled: 32
Confidence: 11.1%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 2 |
|
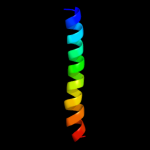
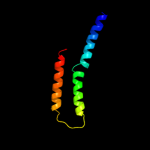
PDB 2rdc chain A
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 9.1%
Identity: 40%
PDB header:lipid binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
Phyre2
| 3 |
|
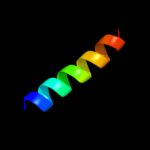
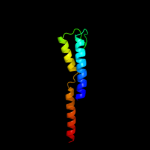
PDB 2l9u chain A
Region: 335 - 354
Aligned: 20
Modelled: 20
Confidence: 7.8%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
Phyre2
| 4 |
|
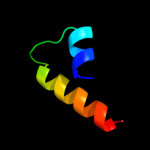
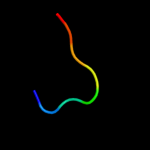
PDB 2voy chain H
Region: 44 - 83
Aligned: 40
Modelled: 40
Confidence: 7.2%
Identity: 10%
PDB header:hydrolase
Chain: H: PDB Molecule:sarcoplasmic/endoplasmic reticulum calcium
PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
Phyre2
| 5 |
|
PDB 1f5q chain B domain 1
Region: 2 - 31
Aligned: 30
Modelled: 30
Confidence: 7.0%
Identity: 3%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 6 |
|
PDB 1mhs chain A
Region: 7 - 83
Aligned: 69
Modelled: 77
Confidence: 7.0%
Identity: 14%
PDB header:membrane protein, proton transport
Chain: A: PDB Molecule:plasma membrane atpase;
PDBTitle: model of neurospora crassa proton atpase
Phyre2
| 7 |
|
PDB 2bbj chain B
Region: 281 - 350
Aligned: 68
Modelled: 70
Confidence: 6.7%
Identity: 16%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 8 |
|
PDB 2zxe chain A
Region: 8 - 83
Aligned: 68
Modelled: 76
Confidence: 5.8%
Identity: 13%
PDB header:hydrolase/transport protein
Chain: A: PDB Molecule:na, k-atpase alpha subunit;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 9 |
|
PDB 1rut chain X domain 1
Region: 2 - 9
Aligned: 8
Modelled: 8
Confidence: 5.6%
Identity: 38%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 10 |
|
PDB 1oed chain A
Region: 6 - 125
Aligned: 120
Modelled: 120
Confidence: 5.2%
Identity: 8%
Fold: Neurotransmitter-gated ion-channel transmembrane pore
Superfamily: Neurotransmitter-gated ion-channel transmembrane pore
Family: Neurotransmitter-gated ion-channel transmembrane pore
Phyre2
| 11 |
|
PDB 2kr6 chain A
Region: 307 - 357
Aligned: 51
Modelled: 51
Confidence: 5.2%
Identity: 20%
PDB header:hydrolase
Chain: A: PDB Molecule:presenilin-1;
PDBTitle: solution structure of presenilin-1 ctf subunit
Phyre2