| 1 | d1v29b_ |
|
 |
24.8 |
13 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Nitrile hydratase beta chain |
| 2 | d1xg8a_ |
|
 |
22.7 |
30 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:YuzD-like |
| 3 | d2pofa1 |
|
 |
22.0 |
33 |
Fold:HIT-like
Superfamily:HIT-like
Family:CDH-like |
| 4 | d1ugpb_ |
|
 |
20.0 |
35 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Nitrile hydratase beta chain |
| 5 | c3dlrA_ |
|
 |
18.5 |
42 |
PDB header:transferase
Chain: A: PDB Molecule:integrase;
PDBTitle: crystal structure of the catalytic core domain from pfv2 integrase
|
| 6 | c2x5cB_ |
|
 |
13.5 |
75 |
PDB header:viral protein
Chain: B: PDB Molecule:hypothetical protein orf131;
PDBTitle: crystal structure of hypothetical protein orf131 from2 pyrobaculum spherical virus
|
| 7 | d2qrda1 |
|
 |
12.2 |
50 |
Fold:TBP-like
Superfamily:KA1-like
Family:Ssp2 C-terminal domain-like |
| 8 | c3qz9D_ |
|
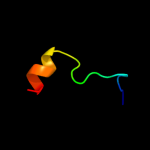 |
12.1 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:co-type nitrile hydratase beta subunit;
PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
|
| 9 | c3l2tB_ |
|
 |
9.5 |
42 |
PDB header:recombination/dna
Chain: B: PDB Molecule:integrase;
PDBTitle: crystal structure of the prototype foamy virus (pfv) intasome in2 complex with magnesium and mk0518 (raltegravir)
|
| 10 | d1c0ma2 |
|
 |
8.8 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 11 | d2ipqx1 |
|
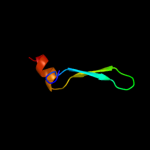 |
7.7 |
27 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:STY4665 C-terminal domain-like |
| 12 | c2de0X_ |
|
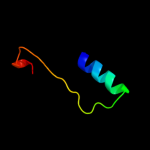 |
7.6 |
19 |
PDB header:transferase
Chain: X: PDB Molecule:alpha-(1,6)-fucosyltransferase;
PDBTitle: crystal structure of human alpha 1,6-fucosyltransferase, fut8
|
| 13 | d1u5ta1 |
|
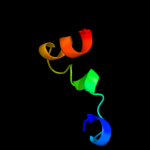 |
6.9 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 14 | c2x5fB_ |
|
 |
6.6 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:aspartate_tyrosine_phenylalanine pyridoxal-5'
PDBTitle: crystal structure of the methicillin-resistant2 staphylococcus aureus sar2028, an3 aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate4 dependent aminotransferase
|
| 15 | d1cxqa_ |
|
 |
6.4 |
8 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 16 | c2b9sA_ |
|
 |
6.3 |
27 |
PDB header:isomerase/dna
Chain: A: PDB Molecule:topoisomerase i-like protein;
PDBTitle: crystal structure of heterodimeric l. donovani2 topoisomerase i-vanadate-dna complex
|
| 17 | c3f1f4_ |
|
 |
5.8 |
67 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: crystal structure of a translation termination complex2 formed with release factor rf2. this file contains the 50s3 subunit of one 70s ribosome. the entire crystal structure4 contains two 70s ribosomes as described in remark 400.
|
| 18 | c2wh44_ |
|
 |
5.3 |
67 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome
|

