| 1 |
|
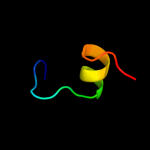
PDB 1y6u chain A
Region: 28 - 46
Aligned: 19
Modelled: 19
Confidence: 50.0%
Identity: 42%
PDB header:dna binding protein
Chain: A: PDB Molecule:excisionase from transposon tn916;
PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
Phyre2
| 2 |
|
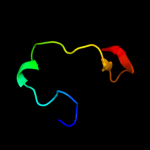
PDB 2lcy chain A
Region: 11 - 19
Aligned: 9
Modelled: 9
Confidence: 10.9%
Identity: 33%
PDB header:viral protein
Chain: A: PDB Molecule:virion spike glycoprotein;
PDBTitle: nmr structure of the complete internal fusion loop from ebolavirus gp22 at ph 5.5
Phyre2
| 3 |
|
PDB 2a5y chain B domain 3
Region: 6 - 43
Aligned: 38
Modelled: 38
Confidence: 9.1%
Identity: 37%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Extended AAA-ATPase domain
Phyre2
| 4 |
|
PDB 3l51 chain A
Region: 19 - 32
Aligned: 14
Modelled: 14
Confidence: 9.0%
Identity: 29%
PDB header:cell cycle
Chain: A: PDB Molecule:structural maintenance of chromosomes protein 2;
PDBTitle: crystal structure of the mouse condensin hinge domain
Phyre2
| 5 |
|
PDB 1f3u chain D
Region: 12 - 39
Aligned: 21
Modelled: 28
Confidence: 7.9%
Identity: 38%
Fold: triple barrel
Superfamily: Rap30/74 interaction domains
Family: Rap30/74 interaction domains
Phyre2
| 6 |
|
PDB 2j8q chain B
Region: 13 - 42
Aligned: 30
Modelled: 30
Confidence: 7.1%
Identity: 23%
PDB header:nuclear protein
Chain: B: PDB Molecule:cleavage and polyadenylation specificity factor 5;
PDBTitle: crystal structure of human cleavage and polyadenylation2 specificity factor 5 (cpsf5) in complex with a sulphate3 ion.
Phyre2
| 7 |
|
PDB 3s88 chain J
Region: 11 - 19
Aligned: 9
Modelled: 9
Confidence: 6.8%
Identity: 44%
PDB header:immune system/viral protein
Chain: J: PDB Molecule:envelope glycoprotein;
PDBTitle: crystal structure of sudan ebolavirus glycoprotein (strain gulu) bound2 to 16f6
Phyre2
| 8 |
|
PDB 2w27 chain A
Region: 14 - 40
Aligned: 27
Modelled: 27
Confidence: 6.2%
Identity: 33%
PDB header:signaling protein
Chain: A: PDB Molecule:ykui protein;
PDBTitle: crystal structure of the bacillus subtilis ykui protein,2 with an eal domain, in complex with substrate c-di-gmp and3 calcium
Phyre2