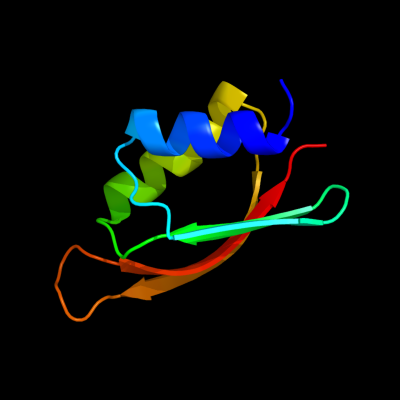
1 c1uv7A_
99.9
36
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
2 d1uv7a_
99.9
36
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM3 c2rjzA_
97.7
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pilo protein;PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
4 c2rddB_
60.1
23
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
5 d1v54d_
53.7
24
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit IVFamily: Mitochondrial cytochrome c oxidase subunit IV6 c2y69Q_
49.0
24
PDB header: electron transportChain: Q: PDB Molecule: cytochrome c oxidase subunit 4 isoform 1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
7 d1yioa1
44.6
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)8 d1ppjc2
24.0
3
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)9 d1t9ma_
23.4
9
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like10 c2y9kG_
20.3
15
PDB header: protein transportChain: G: PDB Molecule: protein invg;PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
11 d1gt0d_
19.2
22
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box12 d1bjna_
18.9
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like13 d1x4pa1
18.3
36
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)14 d1ulra_
17.4
17
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like15 d1k99a_
17.0
13
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box16 c2co9A_
16.6
13
PDB header: transcriptionChain: A: PDB Molecule: thymus high mobility group box protein tox;PDBTitle: solution structure of the hmg_box domain of thymus high2 mobility group box protein tox from mouse
17 d1ty9a_
16.1
11
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: PNP-oxidase like18 d1bccc3
15.0
6
Fold: Heme-binding four-helical bundleSuperfamily: Transmembrane di-heme cytochromesFamily: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)19 c2jsxA_
14.0
7
PDB header: chaperoneChain: A: PDB Molecule: protein napd;PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
20 c2yulA_
13.3
19
PDB header: transcriptionChain: A: PDB Molecule: transcription factor sox-17;PDBTitle: solution structure of the hmg box of human transcription2 factor sox-17
21 c2eefA_
not modelled
12.2
24
PDB header: sugar binding proteinChain: A: PDB Molecule: protein phosphatase 1, regulatory (inhibitor)PDBTitle: solution structure of the cbm_21 domain from human protein2 phosphatase 1, regulatory (inhibitor) subunit 3b
22 d1hywa_
not modelled
11.4
16
Fold: gpW/XkdW-likeSuperfamily: Head-to-tail joining protein W, gpWFamily: Head-to-tail joining protein W, gpW23 c2rnjA_
not modelled
11.3
25
PDB header: transcriptionChain: A: PDB Molecule: response regulator protein vrar;PDBTitle: nmr structure of the s. aureus vrar dna binding domain
24 c2cs1A_
not modelled
11.2
19
PDB header: dna binding proteinChain: A: PDB Molecule: pms1 protein homolog 1;PDBTitle: solution structure of the hmg domain of human dna mismatch2 repair protein
25 c3cwbC_
not modelled
11.1
6
PDB header: oxidoreductaseChain: C: PDB Molecule: cytochrome b;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
26 c2kncA_
not modelled
11.1
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
27 c3mn7S_
not modelled
10.7
50
PDB header: contractile protein/protein bindingChain: S: PDB Molecule: spire ddd;PDBTitle: structures of actin-bound wh2 domains of spire and the implication for2 filament nucleation
28 c3mn5S_
not modelled
10.4
50
PDB header: contractile protein/protein bindingChain: S: PDB Molecule: protein spire;PDBTitle: structures of actin-bound wh2 domains of spire and the implication for2 filament nucleation
29 d1wgfa_
not modelled
10.3
22
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box30 c1x3uA_
not modelled
10.2
35
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein fixj;PDBTitle: solution structure of the c-terminal transcriptional2 activator domain of fixj from sinorhizobium melilot
31 c3gr5A_
not modelled
10.1
15
PDB header: membrane proteinChain: A: PDB Molecule: escc;PDBTitle: periplasmic domain of the outer membrane secretin escc from2 enteropathogenic e.coli (epec)
32 c2k4yA_
not modelled
9.9
6
PDB header: metal transportChain: A: PDB Molecule: feoa-like protein;PDBTitle: nmr structure of feoa-like protein from clostridium2 acetobutylicum: northeast structural genomics consortium3 target car178
33 c1wz6A_
not modelled
9.8
31
PDB header: transcriptionChain: A: PDB Molecule: hmg-box transcription factor bbx;PDBTitle: solution structure of the hmg_box domain of murine bobby2 sox homolog
34 d1w2ia_
not modelled
9.8
13
Fold: Ferredoxin-likeSuperfamily: Acylphosphatase/BLUF domain-likeFamily: Acylphosphatase-like35 d1b7yb4
not modelled
9.8
13
Fold: Ferredoxin-likeSuperfamily: Anticodon-binding domain of PheRSFamily: Anticodon-binding domain of PheRS36 d2c0ra1
not modelled
9.0
6
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like37 c2w7vB_
not modelled
7.7
13
PDB header: transport proteinChain: B: PDB Molecule: general secretion pathway protein l;PDBTitle: periplasmic domain of epsl from vibrio parahaemolyticus
38 c1peiA_
not modelled
7.7
36
PDB header: nucleotidyltransferaseChain: A: PDB Molecule: pepc22;PDBTitle: nmr structure of the membrane-binding domain of ctp2 phosphocholine cytidylyltransferase, 10 structures
39 d1j46a_
not modelled
7.7
24
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box40 c3br8A_
not modelled
7.5
14
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
41 d2af7a1
not modelled
7.5
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like42 d1j3da_
not modelled
7.4
16
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box43 d1l3la1
not modelled
7.2
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: GerE-like (LuxR/UhpA family of transcriptional regulators)44 c2lhjA_
not modelled
7.0
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: high mobility group protein homolog nhp1;PDBTitle: nmr structure of the high mobility group protein-like protein nhp12 from babesia bovis t2bo (baboa.00841.a)
45 c2xlkB_
not modelled
6.8
24
PDB header: hydrolase/rnaChain: B: PDB Molecule: csy4 endoribonuclease;PDBTitle: crystal structure of the csy4-crrna complex, orthorhombic form
46 c2gv1A_
not modelled
6.7
11
PDB header: hydrolaseChain: A: PDB Molecule: probable acylphosphatase;PDBTitle: nmr solution structure of the acylphosphatase from2 eschaerichia coli
47 c3qm2A_
not modelled
6.6
7
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase;PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
48 c2bjeA_
not modelled
6.5
19
PDB header: hydrolaseChain: A: PDB Molecule: acylphosphatase;PDBTitle: acylphosphatase from sulfolobus solfataricus. monclinic p212 space group
49 d1dwka2
not modelled
6.5
24
Fold: Cyanase C-terminal domainSuperfamily: Cyanase C-terminal domainFamily: Cyanase C-terminal domain50 c2k1aA_
not modelled
6.4
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
51 c2iv1J_
not modelled
6.3
16
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
52 d1j5wa_
not modelled
6.1
25
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain53 d1jjcb4
not modelled
6.1
13
Fold: Ferredoxin-likeSuperfamily: Anticodon-binding domain of PheRSFamily: Anticodon-binding domain of PheRS54 c2crjA_
not modelled
6.0
24
PDB header: gene regulationChain: A: PDB Molecule: swi/snf-related matrix-associated actin-PDBTitle: solution structure of the hmg domain of mouse hmg domain2 protein hmgx2
55 c2d88A_
not modelled
6.0
27
PDB header: signaling protein, protein bindingChain: A: PDB Molecule: protein mical-3;PDBTitle: solution structure of the ch domain from human mical-32 protein
56 d1hsma_
not modelled
6.0
13
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box57 c2d89A_
not modelled
5.6
23
PDB header: structural protein, protein bindingChain: A: PDB Molecule: ehbp1 protein;PDBTitle: solution structure of the ch domain from human eh domain2 binding protein 1
58 d2lefa_
not modelled
5.5
19
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box59 d2h3ja1
not modelled
5.4
13
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: FeoA-like60 c2kncB_
not modelled
5.3
19
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
61 c1wylA_
not modelled
5.1
24
PDB header: signaling proteinChain: A: PDB Molecule: nedd9 interacting protein with calponin homologyPDBTitle: solution structure of the ch domain of human nedd92 interacting protein with calponin homology and lim domains
62 c2e6oA_
not modelled
5.1
13
PDB header: transcription, cell cycleChain: A: PDB Molecule: hmg box-containing protein 1;PDBTitle: solution structure of the hmg box domain from human hmg-box2 transcription factor 1