| 1 |
|
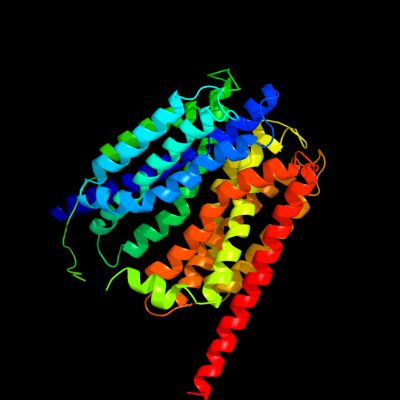
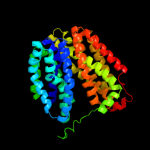
PDB 1pw4 chain A
Region: 2 - 408
Aligned: 406
Modelled: 407
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
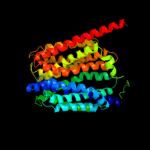
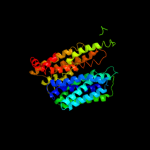
PDB 2gfp chain A
Region: 15 - 388
Aligned: 369
Modelled: 374
Confidence: 100.0%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 3 |
|
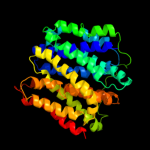
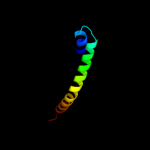
PDB 3o7p chain A
Region: 16 - 397
Aligned: 375
Modelled: 382
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
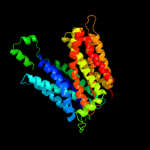
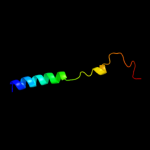
PDB 1pv7 chain A
Region: 16 - 410
Aligned: 395
Modelled: 395
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 15 - 394
Aligned: 380
Modelled: 380
Confidence: 100.0%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 361 - 410
Aligned: 50
Modelled: 50
Confidence: 47.7%
Identity: 16%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 2knc chain A
Region: 374 - 410
Aligned: 37
Modelled: 37
Confidence: 28.8%
Identity: 19%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 8 |
|
PDB 2g9p chain A
Region: 67 - 80
Aligned: 14
Modelled: 14
Confidence: 20.3%
Identity: 36%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 9 |
|
PDB 2xq2 chain A
Region: 276 - 410
Aligned: 129
Modelled: 135
Confidence: 14.9%
Identity: 8%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 10 |
|
PDB 2jln chain A
Region: 355 - 410
Aligned: 56
Modelled: 56
Confidence: 10.2%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 11 |
|
PDB 2rdd chain B
Region: 380 - 408
Aligned: 29
Modelled: 29
Confidence: 7.4%
Identity: 14%
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
Phyre2