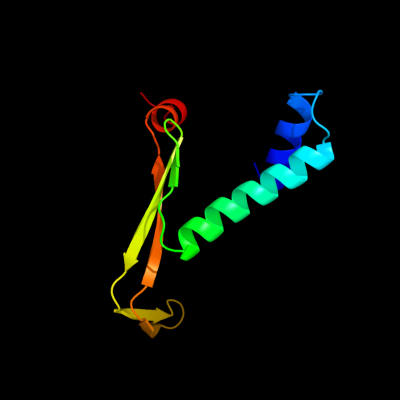
1 d1owfb_
99.9
99
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein2 c2iifA_
99.9
33
PDB header: recombination/dnaChain: A: PDB Molecule: integration host factor;PDBTitle: single chain integration host factor mutant protein (scihf2-2 k45ae) in complex with dna
3 d1p71a_
99.9
33
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein4 c3c4iA_
99.9
33
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein hu homolog;PDBTitle: crystal structure analysis of n terminal region containing the2 dimerization domain and dna binding domain of hu protein(histone like3 protein-dna binding) from mycobacterium tuberculosis [h37rv]
5 d1owfa_
99.9
32
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein6 c2np2B_
99.9
32
PDB header: dna binding protein/dnaChain: B: PDB Molecule: hbb;PDBTitle: hbb-dna complex
7 d1exea_
99.9
28
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein8 d1huua_
99.8
38
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein9 d1mula_
99.7
29
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein10 d2o97b1
99.7
32
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein11 d1b8za_
99.6
45
Fold: IHF-like DNA-binding proteinsSuperfamily: IHF-like DNA-binding proteinsFamily: Prokaryotic DNA-bending protein12 d1i27a_
43.3
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of the rap74 subunit of TFIIF13 d1dula_
35.6
23
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain14 d1o17a1
33.3
20
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain15 c1nriA_
31.5
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein hi0754;PDBTitle: crystal structure of putative phosphosugar isomerase hi0754 from2 haemophilus influenzae
16 d1nria_
31.5
16
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain17 d1nh2d1
29.9
16
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain18 d1hq1a_
29.7
26
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain19 d1brwa1
28.5
17
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain20 d1qzxa2
28.1
29
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain21 c3cvjB_
not modelled
27.4
15
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of a putative phosphoheptose isomerase (bh3325) from2 bacillus halodurans c-125 at 2.00 a resolution
22 c2jqeA_
not modelled
25.2
32
PDB header: signaling proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: soution structure of af54 m-domain
23 d2ffha2
not modelled
21.3
32
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain24 c1o17A_
not modelled
19.3
20
PDB header: transferaseChain: A: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
25 d1khda1
not modelled
18.1
10
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain26 c3tsuA_
not modelled
17.3
30
PDB header: transferaseChain: A: PDB Molecule: transcriptional regulatory protein;PDBTitle: crystal structure of e. coli hypf with amp-pnp and carbamoyl phosphate
27 d1nvpd1
not modelled
16.8
18
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain28 c2k6sB_
not modelled
13.9
16
PDB header: protein transportChain: B: PDB Molecule: rab11fip2 protein;PDBTitle: structure of rab11-fip2 c-terminal coiled-coil domain
29 d2tpta1
not modelled
12.9
7
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain30 d1njra_
not modelled
12.3
11
Fold: Macro domain-likeSuperfamily: Macro domain-likeFamily: Macro domain31 d2o4ta1
not modelled
12.2
15
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like32 d1vola2
not modelled
11.9
15
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain33 d1ussa_
not modelled
11.3
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Linker histone H1/H534 c3kfoA_
not modelled
10.9
9
PDB header: protein transportChain: A: PDB Molecule: nucleoporin nup133;PDBTitle: crystal structure of the c-terminal domain from the nuclear pore2 complex component nup133 from saccharomyces cerevisiae
35 d1ee8a1
not modelled
10.7
11
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins36 c3thgA_
not modelled
10.3
11
PDB header: protein bindingChain: A: PDB Molecule: ribulose bisphosphate carboxylase/oxygenase activase 1,PDBTitle: crystal structure of the creosote rubisco activase c-domain
37 d1uoua1
not modelled
10.2
15
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain38 c2yskA_
not modelled
10.1
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1432;PDBTitle: crystal structure of a hypothetical protein ttha1432 from thermus2 thermophilus
39 d1tdza1
not modelled
9.7
7
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins40 d1dpua_
not modelled
9.3
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of RPA3241 c1dpuA_
not modelled
9.3
20
PDB header: dna binding proteinChain: A: PDB Molecule: replication protein a (rpa32) c-terminal domain;PDBTitle: solution structure of the c-terminal domain of human rpa322 complexed with ung2(73-88)
42 d1soua_
not modelled
9.1
16
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: Methenyltetrahydrofolate synthetase43 d1k82a1
not modelled
9.0
14
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins44 d1z67a1
not modelled
8.9
13
Fold: YidB-likeSuperfamily: YidB-likeFamily: YidB-like45 c2v3cC_
not modelled
8.9
29
PDB header: signaling proteinChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
46 c2vfyA_
not modelled
8.6
22
PDB header: hydrolaseChain: A: PDB Molecule: akap18 delta;PDBTitle: akap18 delta central domain
47 d1qb2a_
not modelled
8.5
20
Fold: Signal peptide-binding domainSuperfamily: Signal peptide-binding domainFamily: Signal peptide-binding domain48 d1r2za1
not modelled
8.2
19
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins49 d1x94a_
not modelled
8.2
21
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain50 c2kloA_
not modelled
8.1
14
PDB header: cell cycleChain: A: PDB Molecule: dna replication factor cdt1;PDBTitle: structure of the cdt1 c-terminal domain
51 d1tk9a_
not modelled
8.0
28
Fold: SIS domainSuperfamily: SIS domainFamily: mono-SIS domain52 c1khdD_
not modelled
7.8
10
PDB header: transferaseChain: D: PDB Molecule: anthranilate phosphoribosyltransferase;PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.93 resolution (current name, pectobacterium carotovorum)
53 c3g2bA_
not modelled
7.8
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: coenzyme pqq synthesis protein d;PDBTitle: crystal structure of pqqd from xanthomonas campestris
54 d1k3xa1
not modelled
7.6
15
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Middle domain of MutM-like DNA repair proteins55 c1nvpD_
not modelled
7.5
18
PDB header: transcription/dnaChain: D: PDB Molecule: transcription initiation factor iia gamma chain;PDBTitle: human tfiia/tbp/dna complex
56 d2fqla1
not modelled
7.5
22
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like57 c1otpA_
not modelled
7.4
7
PDB header: phosphorylaseChain: A: PDB Molecule: thymidine phosphorylase;PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
58 c1tr8A_
not modelled
7.2
21
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
59 c2i2aA_
not modelled
7.1
12
PDB header: transferaseChain: A: PDB Molecule: probable inorganic polyphosphate/atp-nad kinase 1;PDBTitle: crystal structure of lmnadk1 from listeria monocytogenes
60 c3dm5A_
not modelled
6.8
35
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling2 the ribonucleic core of the signal recognition particle3 from the archaeon pyrococcus furiosus.
61 d1aipc1
not modelled
6.5
17
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain62 d1hh2p4
not modelled
6.5
19
Fold: Transcription factor NusA, N-terminal domainSuperfamily: Transcription factor NusA, N-terminal domainFamily: Transcription factor NusA, N-terminal domain63 d1mzba_
not modelled
6.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: FUR-like64 d1aisb2
not modelled
6.2
16
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain65 d2cp9a1
not modelled
6.2
11
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain66 c3eyyA_
not modelled
6.1
19
PDB header: transportChain: A: PDB Molecule: putative iron uptake regulatory protein;PDBTitle: structural basis for the specialization of nur, a nickel-2 specific fur homologue, in metal sensing and dna3 recognition
67 d1z6ra1
not modelled
6.0
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ROK associated domain68 c3jx9B_
not modelled
5.9
19
PDB header: isomeraseChain: B: PDB Molecule: putative phosphoheptose isomerase;PDBTitle: crystal structure of putative phosphoheptose isomerase2 (yp_001815198.1) from exiguobacterium sp. 255-15 at 1.95 a resolution
69 c2az1B_
not modelled
5.8
17
PDB header: transferaseChain: B: PDB Molecule: nucleoside diphosphate kinase;PDBTitle: structure of a halophilic nucleoside diphosphate kinase2 from halobacterium salinarum
70 c3a4cA_
not modelled
5.8
14
PDB header: cell cycle, replicationChain: A: PDB Molecule: dna replication factor cdt1;PDBTitle: crystal structure of cdt1 c terminal domain
71 c2d4gA_
not modelled
5.7
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bsu11850;PDBTitle: structure of yjcg protein, a putative 2'-5' rna ligase from2 bacillus subtilis
72 c2o03A_
not modelled
5.7
23
PDB header: gene regulationChain: A: PDB Molecule: probable zinc uptake regulation protein furb;PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
73 c2dsjA_
not modelled
5.6
13
PDB header: transferaseChain: A: PDB Molecule: pyrimidine-nucleoside (thymidine) phosphorylase;PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
74 c2fu4B_
not modelled
5.5
19
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
75 c1brwB_
not modelled
5.4
17
PDB header: transferaseChain: B: PDB Molecule: protein (pyrimidine nucleoside phosphorylase);PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
76 d1krha1
not modelled
5.4
35
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like77 c3mwmA_
not modelled
5.3
16
PDB header: transcriptionChain: A: PDB Molecule: putative metal uptake regulation protein;PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
78 d2elca1
not modelled
5.3
0
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain79 c2xigA_
not modelled
5.3
6
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
80 d1v8ga1
not modelled
5.3
0
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain81 c1vquB_
not modelled
5.3
10
PDB header: transferaseChain: B: PDB Molecule: anthranilate phosphoribosyltransferase 2;PDBTitle: crystal structure of anthranilate phosphoribosyltransferase 22 (17130499) from nostoc sp. at 1.85 a resolution
82 c3mzyA_
not modelled
5.2
11
PDB header: rna binding proteinChain: A: PDB Molecule: rna polymerase sigma-h factor;PDBTitle: the crystal structure of the rna polymerase sigma-h factor from2 fusobacterium nucleatum to 2.5a
83 c2w2mP_
not modelled
5.2
33
PDB header: hydrolase/receptorChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: wt pcsk9-deltac bound to wt egf-a of ldlr
84 c2pmwA_
not modelled
5.2
33
PDB header: hydrolaseChain: A: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: the crystal structure of proprotein convertase subtilisin2 kexin type 9 (pcsk9)
85 c2p4eP_
not modelled
5.2
33
PDB header: hydrolaseChain: P: PDB Molecule: proprotein convertase subtilisin/kexin type 9;PDBTitle: crystal structure of pcsk9
86 c2q3vB_
not modelled
5.1
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein at2g34160;PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at2g34160