| 1 |
|
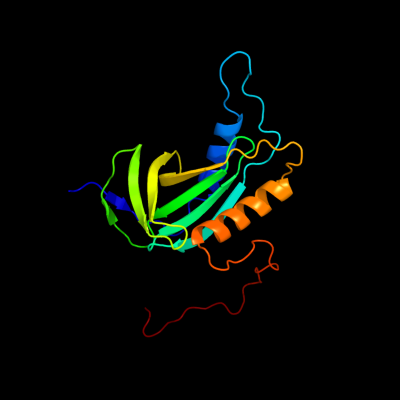
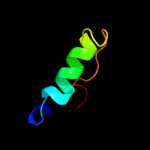
PDB 2kc5 chain A
Region: 1 - 162
Aligned: 162
Modelled: 162
Confidence: 100.0%
Identity: 100%
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase-2 operon protein hybe;
PDBTitle: solution structure of hybe from escherichia coli
Phyre2
| 2 |
|
PDB 1e6v chain A domain 2
Region: 7 - 40
Aligned: 32
Modelled: 34
Confidence: 67.0%
Identity: 16%
Fold: Ferredoxin-like
Superfamily: Methyl-coenzyme M reductase subunits
Family: Methyl-coenzyme M reductase alpha and beta chain N-terminal domain
Phyre2
| 3 |
|
PDB 1hbn chain A domain 2
Region: 7 - 40
Aligned: 32
Modelled: 34
Confidence: 53.8%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: Methyl-coenzyme M reductase subunits
Family: Methyl-coenzyme M reductase alpha and beta chain N-terminal domain
Phyre2
| 4 |
|
PDB 1hbu chain D
Region: 7 - 40
Aligned: 32
Modelled: 34
Confidence: 24.1%
Identity: 19%
PDB header:methanogenesis
Chain: D: PDB Molecule:methyl-coenzyme m reductase i alpha subunit;
PDBTitle: methyl-coenzyme m reductase in the mcr-red1-silent state in2 complex with coenzyme m
Phyre2
| 5 |
|
PDB 1cn3 chain F
Region: 57 - 66
Aligned: 10
Modelled: 10
Confidence: 17.3%
Identity: 50%
PDB header:viral protein
Chain: F: PDB Molecule:fragment of coat protein vp2;
PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
Phyre2
| 6 |
|
PDB 2ide chain E
Region: 31 - 46
Aligned: 15
Modelled: 16
Confidence: 7.4%
Identity: 20%
PDB header:biosynthetic protein
Chain: E: PDB Molecule:molybdenum cofactor biosynthesis protein c;
PDBTitle: crystal structure of the molybdenum cofactor biosynthesis protein c2 (ttha1789) from thermus theromophilus hb8
Phyre2
| 7 |
|
PDB 2eey chain A
Region: 31 - 46
Aligned: 15
Modelled: 16
Confidence: 7.2%
Identity: 20%
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis;
PDBTitle: structure of gk0241 protein from geobacillus kaustophilus
Phyre2
| 8 |
|
PDB 1ekr chain A
Region: 31 - 46
Aligned: 15
Modelled: 16
Confidence: 6.6%
Identity: 13%
Fold: Ferredoxin-like
Superfamily: Molybdenum cofactor biosynthesis protein C, MoaC
Family: Molybdenum cofactor biosynthesis protein C, MoaC
Phyre2
| 9 |
|
PDB 3cmq chain A
Region: 32 - 63
Aligned: 28
Modelled: 32
Confidence: 6.5%
Identity: 29%
PDB header:ligase
Chain: A: PDB Molecule:phenylalanyl-trna synthetase, mitochondrial;
PDBTitle: crystal structure of human mitochondrial phenylalanine trna2 synthetase
Phyre2