| 1 |
|
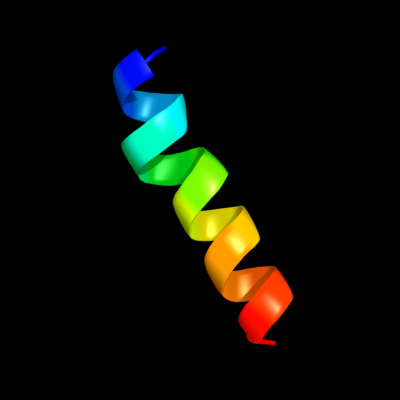
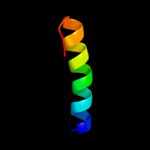
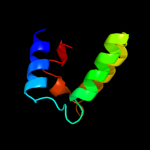
PDB 1v54 chain G
Region: 17 - 35
Aligned: 19
Modelled: 19
Confidence: 19.9%
Identity: 26%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIa
Family: Mitochondrial cytochrome c oxidase subunit VIa
Phyre2
| 2 |
|
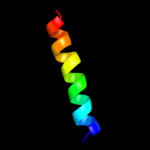
PDB 2gv1 chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 17.6%
Identity: 63%
PDB header:hydrolase
Chain: A: PDB Molecule:probable acylphosphatase;
PDBTitle: nmr solution structure of the acylphosphatase from2 eschaerichia coli
Phyre2
| 3 |
|
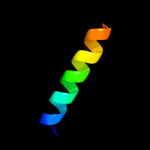
PDB 1ulr chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 15.4%
Identity: 50%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 4 |
|
PDB 3br8 chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 15.1%
Identity: 63%
PDB header:hydrolase
Chain: A: PDB Molecule:probable acylphosphatase;
PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
Phyre2
| 5 |
|
PDB 1w2i chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 14.9%
Identity: 63%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 6 |
|
PDB 3oak chain C
Region: 1 - 15
Aligned: 15
Modelled: 15
Confidence: 14.1%
Identity: 27%
PDB header:transcription
Chain: C: PDB Molecule:transcription elongation factor spt6;
PDBTitle: crystal structure of a spn1 (iws1)-spt6 complex
Phyre2
| 7 |
|
PDB 1aps chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 12.7%
Identity: 50%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 8 |
|
PDB 2bje chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 12.3%
Identity: 63%
PDB header:hydrolase
Chain: A: PDB Molecule:acylphosphatase;
PDBTitle: acylphosphatase from sulfolobus solfataricus. monclinic p212 space group
Phyre2
| 9 |
|
PDB 1urr chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 11.8%
Identity: 50%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 10 |
|
PDB 2acy chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 11.4%
Identity: 50%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 11 |
|
PDB 1gxu chain A
Region: 63 - 70
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 75%
Fold: Ferredoxin-like
Superfamily: Acylphosphatase/BLUF domain-like
Family: Acylphosphatase-like
Phyre2
| 12 |
|
PDB 3nke chain A
Region: 48 - 70
Aligned: 23
Modelled: 23
Confidence: 8.3%
Identity: 22%
PDB header:immune system
Chain: A: PDB Molecule:protein ygbt;
PDBTitle: high resolution structure of the c-terminal domain crisp-associated2 protein cas1 from escherichia coli str. k-12
Phyre2
| 13 |
|
PDB 2yru chain A
Region: 2 - 66
Aligned: 61
Modelled: 65
Confidence: 6.6%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:steroid receptor rna activator 1;
PDBTitle: solution structure of mouse steroid receptor rna activator2 1 (sra1) protein
Phyre2
| 14 |
|
PDB 2k9y chain B
Region: 43 - 64
Aligned: 22
Modelled: 22
Confidence: 6.0%
Identity: 27%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 15 |
|
PDB 3god chain A
Region: 48 - 70
Aligned: 23
Modelled: 23
Confidence: 5.7%
Identity: 22%
PDB header:immune system
Chain: A: PDB Molecule:cas1;
PDBTitle: structural basis for dnase activity of a conserved protein2 implicated in crispr-mediated antiviral defense
Phyre2
| 16 |
|
PDB 1izm chain A
Region: 59 - 67
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 22%
Fold: YgfB-like
Superfamily: YgfB-like
Family: YgfB-like
Phyre2