| 1 |
|
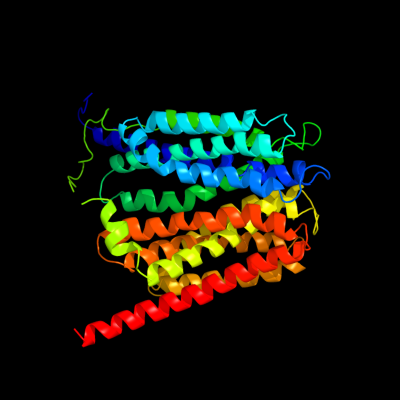
PDB 1pw4 chain A
Region: 3 - 466
Aligned: 415
Modelled: 416
Confidence: 100.0%
Identity: 15%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
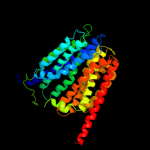
|
PDB 3o7p chain A
Region: 24 - 457
Aligned: 403
Modelled: 409
Confidence: 100.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 3 |
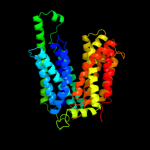
|
PDB 2gfp chain A
Region: 21 - 446
Aligned: 369
Modelled: 370
Confidence: 100.0%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
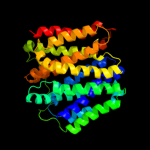
|
PDB 2xut chain C
Region: 23 - 454
Aligned: 432
Modelled: 432
Confidence: 100.0%
Identity: 9%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 5 |
|
PDB 1pv7 chain A
Region: 14 - 470
Aligned: 409
Modelled: 411
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 337 - 495
Aligned: 155
Modelled: 159
Confidence: 70.2%
Identity: 7%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 3hd6 chain A
Region: 338 - 483
Aligned: 146
Modelled: 146
Confidence: 24.6%
Identity: 12%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 8 |
|
PDB 2g9p chain A
Region: 74 - 87
Aligned: 14
Modelled: 14
Confidence: 17.5%
Identity: 21%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 9 |
|
PDB 2knc chain A
Region: 427 - 473
Aligned: 47
Modelled: 47
Confidence: 9.2%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 3b60 chain A domain 2
Region: 11 - 85
Aligned: 75
Modelled: 75
Confidence: 7.3%
Identity: 11%
Fold: ABC transporter transmembrane region
Superfamily: ABC transporter transmembrane region
Family: ABC transporter transmembrane region
Phyre2
| 11 |
|
PDB 3din chain D
Region: 8 - 51
Aligned: 44
Modelled: 44
Confidence: 6.9%
Identity: 9%
PDB header:membrane protein, protein transport
Chain: D: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 12 |
|
PDB 3pro chain C domain 1
Region: 42 - 83
Aligned: 42
Modelled: 42
Confidence: 6.5%
Identity: 12%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 13 |
|
PDB 3mk7 chain F
Region: 433 - 483
Aligned: 51
Modelled: 51
Confidence: 6.4%
Identity: 10%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2