| 1 |
|
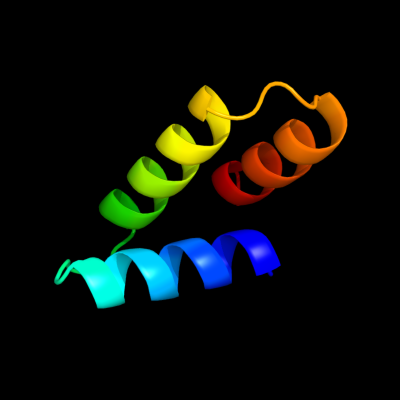
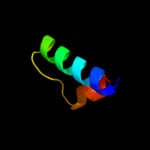
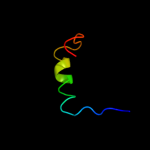
PDB 2oxl chain A
Region: 27 - 75
Aligned: 49
Modelled: 49
Confidence: 97.4%
Identity: 20%
PDB header:gene regulation
Chain: A: PDB Molecule:hypothetical protein ymgb;
PDBTitle: structure and function of the e. coli protein ymgb: a protein critical2 for biofilm formation and acid resistance
Phyre2
| 2 |
|
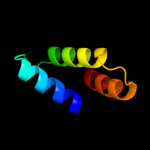
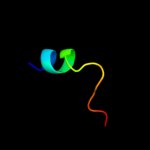
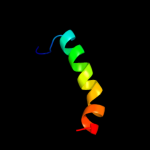
PDB 3c6a chain A
Region: 2 - 58
Aligned: 57
Modelled: 57
Confidence: 41.0%
Identity: 23%
PDB header:viral protein
Chain: A: PDB Molecule:terminase large subunit;
PDBTitle: crystal structure of the rb49 gp17 nuclease domain
Phyre2
| 3 |
|
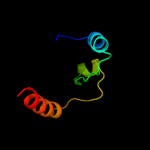
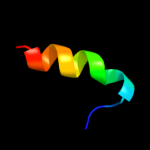
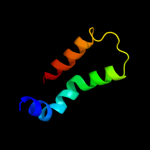
PDB 1stz chain A domain 1
Region: 24 - 53
Aligned: 30
Modelled: 30
Confidence: 36.5%
Identity: 20%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Heat-inducible transcription repressor HrcA, N-terminal domain
Phyre2
| 4 |
|
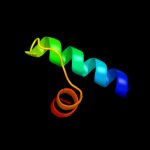
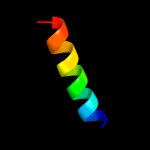
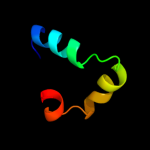
PDB 1j2j chain B
Region: 51 - 73
Aligned: 23
Modelled: 23
Confidence: 27.1%
Identity: 35%
Fold: Spectrin repeat-like
Superfamily: GAT-like domain
Family: GAT domain
Phyre2
| 5 |
|
PDB 2l82 chain A
Region: 21 - 42
Aligned: 22
Modelled: 22
Confidence: 16.7%
Identity: 36%
PDB header:de novo protein
Chain: A: PDB Molecule:designed protein or32;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
Phyre2
| 6 |
|
PDB 1jhf chain A domain 1
Region: 24 - 53
Aligned: 30
Modelled: 30
Confidence: 15.1%
Identity: 27%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: LexA repressor, N-terminal DNA-binding domain
Phyre2
| 7 |
|
PDB 2dzl chain A
Region: 11 - 26
Aligned: 16
Modelled: 16
Confidence: 13.0%
Identity: 19%
PDB header:structural genomics unknown function
Chain: A: PDB Molecule:protein fam100b;
PDBTitle: solution structure of the uba domain in human protein2 fam100b
Phyre2
| 8 |
|
PDB 1bcg chain A
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 9.8%
Identity: 42%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Scorpion toxin-like
Family: Long-chain scorpion toxins
Phyre2
| 9 |
|
PDB 3p3d chain A
Region: 5 - 20
Aligned: 16
Modelled: 16
Confidence: 9.5%
Identity: 19%
PDB header:nuclear protein
Chain: A: PDB Molecule:nucleoporin 53;
PDBTitle: crystal structure of the nup53 rrm domain from pichia guilliermondii
Phyre2
| 10 |
|
PDB 1wwh chain B
Region: 5 - 20
Aligned: 16
Modelled: 16
Confidence: 6.9%
Identity: 31%
PDB header:protein transport
Chain: B: PDB Molecule:nucleoporin 35;
PDBTitle: crystal structure of the mppn domain of mouse nup35
Phyre2
| 11 |
|
PDB 1wwh chain A domain 1
Region: 5 - 20
Aligned: 16
Modelled: 16
Confidence: 6.7%
Identity: 31%
Fold: Ferredoxin-like
Superfamily: RNA-binding domain, RBD
Family: Canonical RBD
Phyre2
| 12 |
|
PDB 3mgj chain A
Region: 27 - 44
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1480;
PDBTitle: crystal structure of the saccharop_dh_n domain of mj14802 protein from methanococcus jannaschii. northeast structural3 genomics consortium target mjr83a.
Phyre2
| 13 |
|
PDB 1x3a chain A domain 1
Region: 15 - 30
Aligned: 16
Modelled: 16
Confidence: 5.7%
Identity: 31%
Fold: BSD domain-like
Superfamily: BSD domain-like
Family: BSD domain
Phyre2
| 14 |
|
PDB 1au7 chain B
Region: 2 - 29
Aligned: 28
Modelled: 28
Confidence: 5.6%
Identity: 18%
PDB header:transcription/dna
Chain: B: PDB Molecule:protein pit-1;
PDBTitle: pit-1 mutant/dna complex
Phyre2
| 15 |
|
PDB 1zav chain A domain 1
Region: 21 - 46
Aligned: 26
Modelled: 26
Confidence: 5.5%
Identity: 19%
Fold: Ferredoxin-like
Superfamily: Ribosomal protein L10-like
Family: Ribosomal protein L10-like
Phyre2
| 16 |
|
PDB 3bxj chain B
Region: 13 - 62
Aligned: 50
Modelled: 50
Confidence: 5.5%
Identity: 20%
PDB header:signaling protein
Chain: B: PDB Molecule:ras gtpase-activating protein syngap;
PDBTitle: crystal structure of the c2-gap fragment of syngap
Phyre2
| 17 |
|
PDB 3idw chain A
Region: 5 - 40
Aligned: 34
Modelled: 36
Confidence: 5.3%
Identity: 18%
PDB header:endocytosis
Chain: A: PDB Molecule:actin cytoskeleton-regulatory complex protein sla1;
PDBTitle: crystal structure of sla1 homology domain 2
Phyre2