| 1 |
|
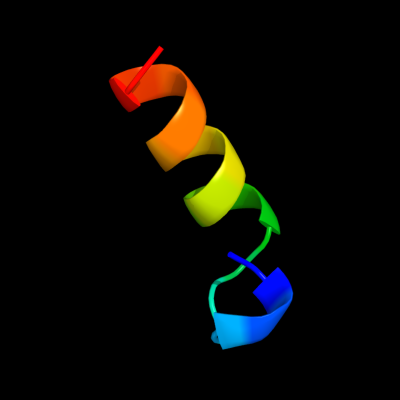
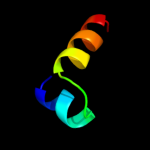
PDB 1z96 chain A domain 1
Region: 146 - 164
Aligned: 19
Modelled: 19
Confidence: 27.7%
Identity: 53%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: UBA domain
Phyre2
| 2 |
|
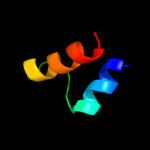
PDB 1cuk chain A domain 1
Region: 146 - 156
Aligned: 11
Modelled: 11
Confidence: 21.4%
Identity: 64%
Fold: RuvA C-terminal domain-like
Superfamily: DNA helicase RuvA subunit, C-terminal domain
Family: DNA helicase RuvA subunit, C-terminal domain
Phyre2
| 3 |
|
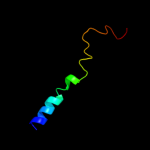
PDB 2axt chain K domain 1
Region: 10 - 39
Aligned: 30
Modelled: 30
Confidence: 18.3%
Identity: 17%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein K, PsbK
Family: PsbK-like
Phyre2
| 4 |
|
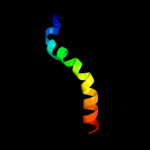
PDB 2lbg chain A
Region: 455 - 470
Aligned: 16
Modelled: 16
Confidence: 18.2%
Identity: 38%
PDB header:membrane protein
Chain: A: PDB Molecule:major prion protein;
PDBTitle: structure of the chr of the prion protein in dpc micelles
Phyre2
| 5 |
|
PDB 3a0b chain K
Region: 10 - 39
Aligned: 30
Modelled: 30
Confidence: 10.8%
Identity: 17%
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 6 |
|
PDB 1y60 chain A
Region: 466 - 486
Aligned: 21
Modelled: 21
Confidence: 8.3%
Identity: 19%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: Formaldehyde-activating enzyme, FAE
Phyre2
| 7 |
|
PDB 3orc chain A
Region: 479 - 500
Aligned: 22
Modelled: 22
Confidence: 8.2%
Identity: 27%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: Phage repressors
Phyre2
| 8 |
|
PDB 2z3x chain C
Region: 477 - 507
Aligned: 31
Modelled: 31
Confidence: 8.1%
Identity: 16%
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:small, acid-soluble spore protein c;
PDBTitle: structure of a protein-dna complex essential for dna2 protection in spore of bacillus species
Phyre2
| 9 |
|
PDB 2knc chain A
Region: 249 - 284
Aligned: 36
Modelled: 36
Confidence: 7.2%
Identity: 14%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 3a0h chain K
Region: 10 - 39
Aligned: 30
Modelled: 30
Confidence: 7.2%
Identity: 17%
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 11 |
|
PDB 3a0b chain K
Region: 10 - 39
Aligned: 30
Modelled: 30
Confidence: 6.8%
Identity: 17%
PDB header:electron transport
Chain: K: PDB Molecule:photosystem ii reaction center protein k;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 12 |
|
PDB 1d1l chain A
Region: 479 - 500
Aligned: 22
Modelled: 22
Confidence: 5.4%
Identity: 27%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: Phage repressors
Phyre2
| 13 |
|
PDB 2i7p chain A domain 1
Region: 455 - 468
Aligned: 14
Modelled: 14
Confidence: 5.4%
Identity: 21%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Fumble-like
Phyre2
| 14 |
|
PDB 2o01 chain F
Region: 447 - 461
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 33%
PDB header:photosynthesis
Chain: F: PDB Molecule:photosystem i reaction center subunit iii,
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 15 |
|
PDB 2pij chain B
Region: 479 - 500
Aligned: 22
Modelled: 22
Confidence: 5.3%
Identity: 23%
PDB header:transcription
Chain: B: PDB Molecule:prophage pfl 6 cro;
PDBTitle: structure of the cro protein from prophage pfl 6 in pseudomonas2 fluorescens pf-5
Phyre2
| 16 |
|
PDB 1jb0 chain F
Region: 447 - 461
Aligned: 15
Modelled: 15
Confidence: 5.2%
Identity: 33%
Fold: Single transmembrane helix
Superfamily: Subunit III of photosystem I reaction centre, PsaF
Family: Subunit III of photosystem I reaction centre, PsaF
Phyre2
| 17 |
|
PDB 2nww chain A domain 1
Region: 522 - 555
Aligned: 34
Modelled: 34
Confidence: 5.2%
Identity: 15%
Fold: Proton glutamate symport protein
Superfamily: Proton glutamate symport protein
Family: Proton glutamate symport protein
Phyre2