| 1 |
|
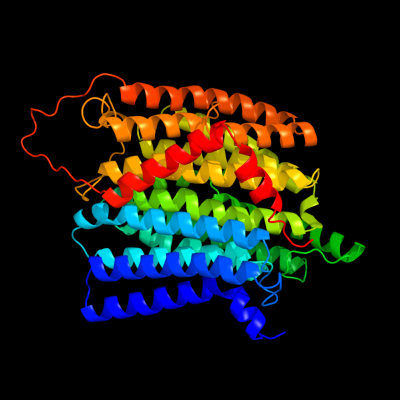
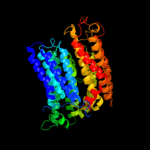
PDB 3rko chain N
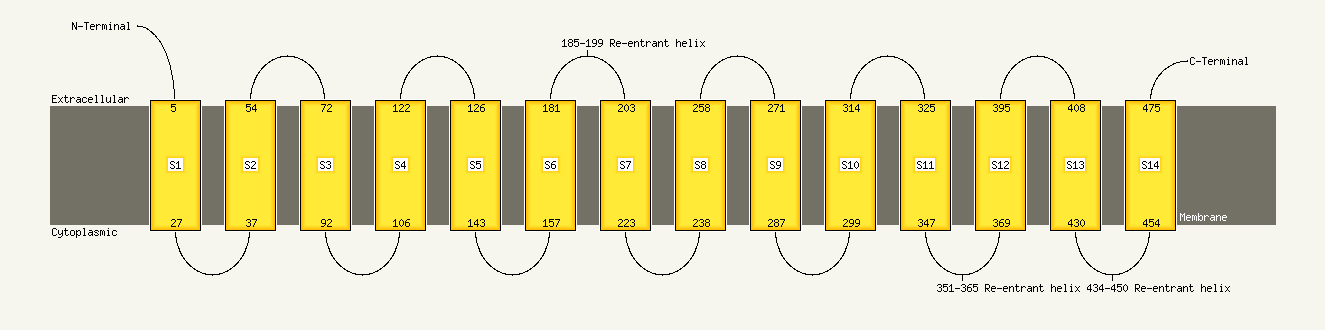
Region: 1 - 485
Aligned: 473
Modelled: 485
Confidence: 100.0%
Identity: 99%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
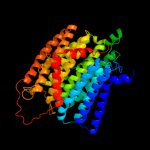
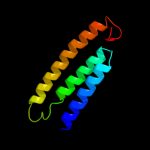
PDB 3rko chain L
Region: 3 - 473
Aligned: 457
Modelled: 471
Confidence: 100.0%
Identity: 19%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
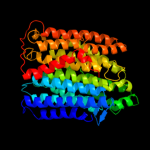
PDB 3rko chain M
Region: 8 - 483
Aligned: 467
Modelled: 476
Confidence: 100.0%
Identity: 20%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 4 |
|
PDB 3rko chain K
Region: 102 - 188
Aligned: 87
Modelled: 87
Confidence: 54.7%
Identity: 21%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 5 |
|
PDB 1a6q chain A domain 1
Region: 371 - 405
Aligned: 35
Modelled: 35
Confidence: 39.8%
Identity: 11%
Fold: Another 3-helical bundle
Superfamily: Protein serine/threonine phosphatase 2C, C-terminal domain
Family: Protein serine/threonine phosphatase 2C, C-terminal domain
Phyre2
| 6 |
|
PDB 2l3i chain A
Region: 288 - 300
Aligned: 13
Modelled: 13
Confidence: 21.6%
Identity: 46%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:aoxki4a, antimicrobial peptide in spider venom;
PDBTitle: oxki4a, spider derived antimicrobial peptide
Phyre2
| 7 |
|
PDB 3mes chain B
Region: 215 - 227
Aligned: 13
Modelled: 13
Confidence: 8.8%
Identity: 15%
PDB header:transferase
Chain: B: PDB Molecule:choline kinase;
PDBTitle: crystal structure of choline kinase from cryptosporidium2 parvum iowa ii, cgd3_2030
Phyre2
| 8 |
|
PDB 1fft chain B domain 2
Region: 402 - 481
Aligned: 80
Modelled: 80
Confidence: 8.0%
Identity: 10%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 9 |
|
PDB 2knc chain A
Region: 404 - 448
Aligned: 45
Modelled: 45
Confidence: 7.2%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 10 |
|
PDB 1r6r chain A
Region: 370 - 396
Aligned: 27
Modelled: 27
Confidence: 6.8%
Identity: 26%
PDB header:viral protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: solution structure of dengue virus capsid protein reveals a2 new fold
Phyre2
| 11 |
|
PDB 1r6r chain A
Region: 370 - 396
Aligned: 27
Modelled: 27
Confidence: 6.8%
Identity: 26%
Fold: Flavivirus capsid protein C
Superfamily: Flavivirus capsid protein C
Family: Flavivirus capsid protein C
Phyre2
| 12 |
|
PDB 1ceu chain A
Region: 222 - 233
Aligned: 12
Modelled: 12
Confidence: 6.6%
Identity: 25%
PDB header:viral protein
Chain: A: PDB Molecule:protein (hiv-1 regulatory protein n-terminal
PDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
Phyre2
| 13 |
|
PDB 1n9w chain A domain 2
Region: 385 - 392
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 50%
Fold: Class II aaRS and biotin synthetases
Superfamily: Class II aaRS and biotin synthetases
Family: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
Phyre2
| 14 |
|
PDB 3r24 chain A
Region: 224 - 232
Aligned: 9
Modelled: 9
Confidence: 6.5%
Identity: 44%
PDB header:transferase, viral protein
Chain: A: PDB Molecule:2'-o-methyl transferase;
PDBTitle: crystal structure of nsp10/nsp16 complex of sars coronavirus" if2 possible
Phyre2
| 15 |
|
PDB 3c25 chain A
Region: 223 - 228
Aligned: 6
Modelled: 6
Confidence: 6.3%
Identity: 67%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:noti restriction endonuclease;
PDBTitle: crystal structure of noti restriction endonuclease bound to cognate2 dna
Phyre2
| 16 |
|
PDB 1n9w chain A
Region: 385 - 392
Aligned: 8
Modelled: 8
Confidence: 6.2%
Identity: 50%
PDB header:biosynthetic protein
Chain: A: PDB Molecule:aspartyl-trna synthetase 2;
PDBTitle: crystal structure of the non-discriminating and archaeal-2 type aspartyl-trna synthetase from thermus thermophilus
Phyre2
| 17 |
|
PDB 3dtu chain B domain 2
Region: 384 - 465
Aligned: 77
Modelled: 82
Confidence: 6.1%
Identity: 9%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 18 |
|
PDB 3bju chain B
Region: 385 - 392
Aligned: 8
Modelled: 8
Confidence: 5.8%
Identity: 50%
PDB header:ligase
Chain: B: PDB Molecule:lysyl-trna synthetase;
PDBTitle: crystal structure of tetrameric form of human lysyl-trna2 synthetase
Phyre2
| 19 |
|
PDB 1b8a chain B
Region: 385 - 392
Aligned: 8
Modelled: 8
Confidence: 5.4%
Identity: 50%
PDB header:ligase
Chain: B: PDB Molecule:protein (aspartyl-trna synthetase);
PDBTitle: aspartyl-trna synthetase
Phyre2
| 20 |
|
PDB 2xte chain H
Region: 469 - 483
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 40%
PDB header:transcription
Chain: H: PDB Molecule:f-box-like/wd repeat-containing protein tbl1x;
PDBTitle: structure of the tbl1 tetramerisation domain
Phyre2
| 21 |
|
| 22 |
|