| 1 |
|
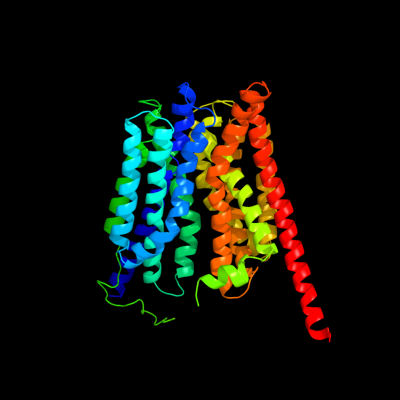
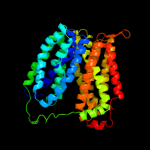
PDB 1pw4 chain A
Region: 1 - 467
Aligned: 412
Modelled: 413
Confidence: 100.0%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
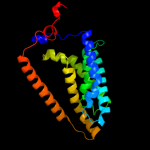
PDB 2gfp chain A
Region: 12 - 442
Aligned: 370
Modelled: 379
Confidence: 100.0%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 3 |
|
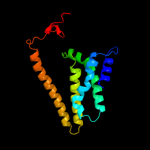
PDB 2xut chain C
Region: 12 - 448
Aligned: 437
Modelled: 437
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 4 |
|
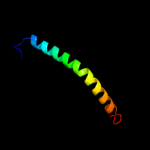
PDB 3o7p chain A
Region: 7 - 453
Aligned: 407
Modelled: 422
Confidence: 100.0%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 1pv7 chain A
Region: 1 - 466
Aligned: 407
Modelled: 424
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 6 |
|
PDB 3b9y chain A
Region: 255 - 475
Aligned: 208
Modelled: 221
Confidence: 85.6%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 7 |
|
PDB 3hd6 chain A
Region: 310 - 475
Aligned: 161
Modelled: 161
Confidence: 64.8%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 8 |
|
PDB 3qnq chain D
Region: 424 - 468
Aligned: 45
Modelled: 45
Confidence: 45.9%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 9 |
|
PDB 3cx5 chain C domain 2
Region: 7 - 67
Aligned: 61
Modelled: 61
Confidence: 14.7%
Identity: 11%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 10 |
|
PDB 2g9p chain A
Region: 65 - 78
Aligned: 14
Modelled: 14
Confidence: 14.1%
Identity: 36%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 11 |
|
PDB 2qjk chain M
Region: 7 - 67
Aligned: 61
Modelled: 61
Confidence: 10.2%
Identity: 11%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 12 |
|
PDB 2oar chain A domain 1
Region: 391 - 475
Aligned: 81
Modelled: 85
Confidence: 8.9%
Identity: 9%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 13 |
|
PDB 3cx5 chain N
Region: 7 - 67
Aligned: 61
Modelled: 61
Confidence: 8.1%
Identity: 11%
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
Phyre2
| 14 |
|
PDB 2jo1 chain A
Region: 432 - 475
Aligned: 38
Modelled: 44
Confidence: 7.6%
Identity: 18%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 15 |
|
PDB 1q90 chain B
Region: 7 - 67
Aligned: 61
Modelled: 61
Confidence: 7.4%
Identity: 8%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 16 |
|
PDB 2knc chain A
Region: 429 - 470
Aligned: 42
Modelled: 42
Confidence: 6.6%
Identity: 5%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2