| 1 |
|
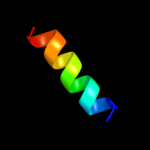
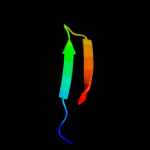
PDB 1zll chain E
Region: 7 - 19
Aligned: 13
Modelled: 13
Confidence: 20.3%
Identity: 31%
PDB header:membrane protein/signaling protein
Chain: E: PDB Molecule:cardiac phospholamban;
PDBTitle: nmr structure of unphosphorylated human phospholamban2 pentamer
Phyre2
| 2 |
|
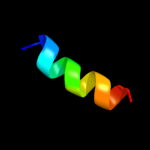
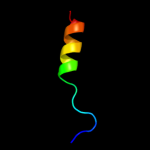
PDB 3arc chain L
Region: 7 - 22
Aligned: 16
Modelled: 16
Confidence: 18.2%
Identity: 19%
PDB header:electron transport, photosynthesis
Chain: L: PDB Molecule:photosystem ii reaction center protein l;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 3 |
|
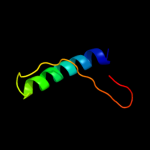
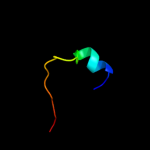
PDB 2wj8 chain N
Region: 9 - 22
Aligned: 14
Modelled: 14
Confidence: 17.0%
Identity: 36%
PDB header:rna binding protein/rna
Chain: N: PDB Molecule:nucleoprotein;
PDBTitle: respiratory syncitial virus ribonucleoprotein
Phyre2
| 4 |
|
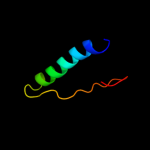
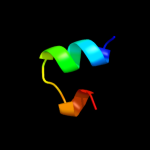
PDB 1y4o chain A domain 1
Region: 3 - 41
Aligned: 39
Modelled: 39
Confidence: 11.8%
Identity: 18%
Fold: Profilin-like
Superfamily: Roadblock/LC7 domain
Family: Roadblock/LC7 domain
Phyre2
| 5 |
|
PDB 2hz5 chain A domain 1
Region: 3 - 41
Aligned: 39
Modelled: 39
Confidence: 9.7%
Identity: 18%
Fold: Profilin-like
Superfamily: Roadblock/LC7 domain
Family: Roadblock/LC7 domain
Phyre2
| 6 |
|
PDB 1prt chain C domain 1
Region: 28 - 43
Aligned: 16
Modelled: 16
Confidence: 8.5%
Identity: 38%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 7 |
|
PDB 1xme chain C domain 1
Region: 2 - 21
Aligned: 20
Modelled: 20
Confidence: 8.0%
Identity: 20%
Fold: Single transmembrane helix
Superfamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
Family: Bacterial ba3 type cytochrome c oxidase subunit IIa
Phyre2
| 8 |
|
PDB 2k4f chain A
Region: 34 - 41
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 50%
PDB header:immune system, signaling protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd3 epsilon chain;
PDBTitle: mouse cd3epsilon cytoplasmic tail
Phyre2
| 9 |
|
PDB 1prt chain B domain 1
Region: 25 - 43
Aligned: 19
Modelled: 19
Confidence: 6.0%
Identity: 37%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Bacterial AB5 toxins, B-subunits
Phyre2
| 10 |
|
PDB 1h9r chain A domain 1
Region: 24 - 46
Aligned: 23
Modelled: 23
Confidence: 5.4%
Identity: 13%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 11 |
|
PDB 2km6 chain A
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 5.4%
Identity: 19%
PDB header:signaling protein, protein binding
Chain: A: PDB Molecule:nacht, lrr and pyd domains-containing protein 7;
PDBTitle: nmr structure of the nlrp7 pyrin domain
Phyre2
| 12 |
|
PDB 1h9m chain A domain 2
Region: 24 - 48
Aligned: 25
Modelled: 25
Confidence: 5.3%
Identity: 28%
Fold: OB-fold
Superfamily: MOP-like
Family: BiMOP, duplicated molybdate-binding domain
Phyre2
| 13 |
|
PDB 1pn5 chain A
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 5.0%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:nacht-, lrr- and pyd-containing protein 2;
PDBTitle: nmr structure of the nalp1 pyrin domain (pyd)
Phyre2