| 1 |
|
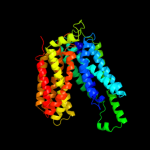
PDB 2xut chain C
Region: 15 - 473
Aligned: 448
Modelled: 459
Confidence: 100.0%
Identity: 17%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 2 |
|
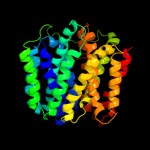
PDB 1pw4 chain A
Region: 2 - 489
Aligned: 414
Modelled: 419
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
PDB 1pv7 chain A
Region: 14 - 487
Aligned: 401
Modelled: 402
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
PDB 3o7p chain A
Region: 17 - 479
Aligned: 408
Modelled: 411
Confidence: 100.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2gfp chain A
Region: 16 - 474
Aligned: 369
Modelled: 372
Confidence: 100.0%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 6 |
|
PDB 3pro chain C domain 1
Region: 35 - 79
Aligned: 45
Modelled: 45
Confidence: 10.9%
Identity: 16%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Alpha-lytic protease prodomain
Family: Alpha-lytic protease prodomain
Phyre2
| 7 |
|
PDB 1jo5 chain A
Region: 452 - 479
Aligned: 28
Modelled: 28
Confidence: 8.7%
Identity: 18%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 8 |
|
PDB 1wrg chain A
Region: 452 - 479
Aligned: 28
Modelled: 28
Confidence: 7.2%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:light-harvesting protein b-880, beta chain;
PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
Phyre2
| 9 |
|
PDB 1o82 chain A
Region: 41 - 74
Aligned: 34
Modelled: 34
Confidence: 5.5%
Identity: 12%
Fold: Saposin-like
Superfamily: Bacteriocin AS-48
Family: Bacteriocin AS-48
Phyre2