| 1 |
|
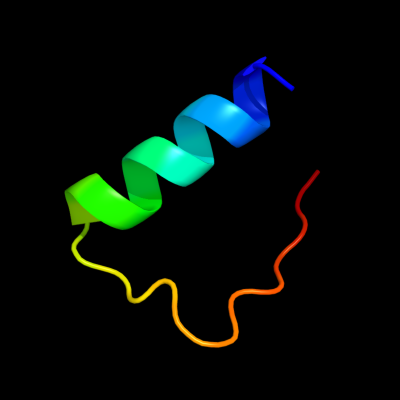
PDB 1h41 chain A domain 2
Region: 4 - 29
Aligned: 26
Modelled: 26
Confidence: 7.3%
Identity: 31%
Fold: Zincin-like
Superfamily: beta-N-acetylhexosaminidase-like domain
Family: alpha-D-glucuronidase, N-terminal domain
Phyre2
| 2 |
|
PDB 1jug chain A
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.6%
Identity: 57%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: C-type lysozyme
Phyre2
| 3 |
|
PDB 1gd6 chain A
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.5%
Identity: 57%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: C-type lysozyme
Phyre2
| 4 |
|
PDB 1n13 chain J
Region: 31 - 66
Aligned: 36
Modelled: 36
Confidence: 5.4%
Identity: 22%
PDB header:lyase
Chain: J: PDB Molecule:pyruvoyl-dependent arginine decarboxylase alpha
PDBTitle: the crystal structure of pyruvoyl-dependent arginine2 decarboxylase from methanococcus jannashii
Phyre2
| 5 |
|
PDB 2vb1 chain A domain 1
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.4%
Identity: 57%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: C-type lysozyme
Phyre2
| 6 |
|
PDB 1yro chain A domain 1
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.3%
Identity: 71%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: C-type lysozyme
Phyre2
| 7 |
|
PDB 1ghl chain A
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.3%
Identity: 57%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: C-type lysozyme
Phyre2
| 8 |
|
PDB 2goi chain C
Region: 6 - 12
Aligned: 7
Modelled: 7
Confidence: 5.2%
Identity: 71%
PDB header:cell adhesion, sugar binding protein
Chain: C: PDB Molecule:sperm lysozyme-like protein 1;
PDBTitle: crystal structure of mouse sperm c-type lysozyme-like2 protein 1
Phyre2