| 1 |
|
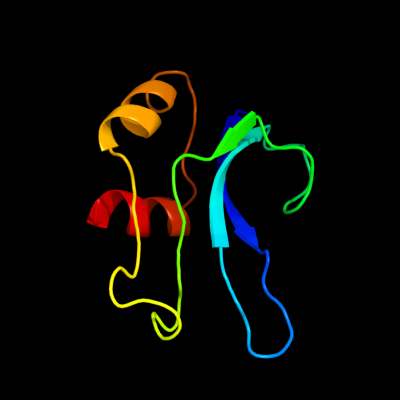
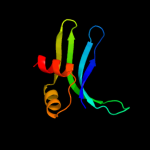
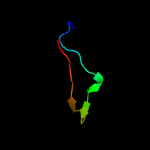
PDB 2gy9 chain P domain 1
Region: 1 - 78
Aligned: 78
Modelled: 78
Confidence: 100.0%
Identity: 100%
Fold: Ribosomal protein S16
Superfamily: Ribosomal protein S16
Family: Ribosomal protein S16
Phyre2
| 2 |
|
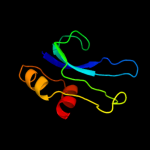
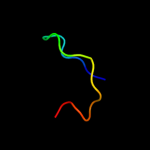
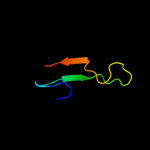
PDB 2uub chain P domain 1
Region: 1 - 79
Aligned: 78
Modelled: 79
Confidence: 100.0%
Identity: 36%
Fold: Ribosomal protein S16
Superfamily: Ribosomal protein S16
Family: Ribosomal protein S16
Phyre2
| 3 |
|
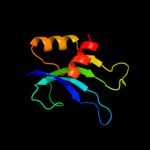
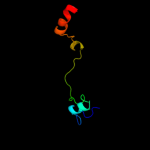
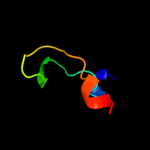
PDB 3bbn chain P
Region: 1 - 79
Aligned: 75
Modelled: 79
Confidence: 100.0%
Identity: 37%
PDB header:ribosome
Chain: P: PDB Molecule:ribosomal protein s16;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
Phyre2
| 4 |
|
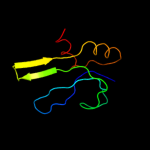
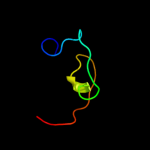
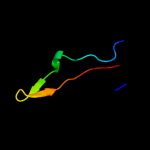
PDB 3bn0 chain A domain 1
Region: 2 - 79
Aligned: 69
Modelled: 78
Confidence: 100.0%
Identity: 29%
Fold: Ribosomal protein S16
Superfamily: Ribosomal protein S16
Family: Ribosomal protein S16
Phyre2
| 5 |
|
PDB 1vbv chain A domain 1
Region: 12 - 27
Aligned: 16
Modelled: 16
Confidence: 48.4%
Identity: 31%
Fold: SH3-like barrel
Superfamily: YccV-like
Family: YccV-like
Phyre2
| 6 |
|
PDB 3f62 chain A
Region: 20 - 37
Aligned: 18
Modelled: 18
Confidence: 25.7%
Identity: 28%
PDB header:cytokine
Chain: A: PDB Molecule:interleukin 18 binding protein;
PDBTitle: crystal structure of human il-18 in complex with ectromelia virus il-2 18 binding protein
Phyre2
| 7 |
|
PDB 3g0k chain A
Region: 1 - 46
Aligned: 44
Modelled: 46
Confidence: 18.6%
Identity: 11%
PDB header:ca-binding protein
Chain: A: PDB Molecule:putative membrane protein;
PDBTitle: crystal structure of a protein of unknown function with a cystatin-2 like fold (saro_2880) from novosphingobium aromaticivorans dsm at3 1.30 a resolution
Phyre2
| 8 |
|
PDB 1vqo chain L domain 1
Region: 7 - 64
Aligned: 46
Modelled: 58
Confidence: 10.2%
Identity: 28%
Fold: Ribosomal proteins L15p and L18e
Superfamily: Ribosomal proteins L15p and L18e
Family: Ribosomal proteins L15p and L18e
Phyre2
| 9 |
|
PDB 2heq chain A domain 1
Region: 7 - 36
Aligned: 19
Modelled: 30
Confidence: 10.0%
Identity: 37%
Fold: SH3-like barrel
Superfamily: YorP-like
Family: YorP-like
Phyre2
| 10 |
|
PDB 3dkx chain A
Region: 10 - 22
Aligned: 13
Modelled: 13
Confidence: 9.7%
Identity: 23%
PDB header:replication
Chain: A: PDB Molecule:replication protein repb;
PDBTitle: crystal structure of the replication initiator protein2 encoded on plasmid pmv158 (repb), trigonal form, to 2.7 ang3 resolution
Phyre2
| 11 |
|
PDB 3cmc chain O domain 2
Region: 13 - 40
Aligned: 28
Modelled: 28
Confidence: 9.1%
Identity: 14%
Fold: FwdE/GAPDH domain-like
Superfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family: GAPDH-like
Phyre2
| 12 |
|
PDB 2dlx chain A domain 1
Region: 6 - 41
Aligned: 31
Modelled: 36
Confidence: 8.8%
Identity: 10%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: UAS domain
Phyre2
| 13 |
|
PDB 2v3m chain F
Region: 9 - 19
Aligned: 11
Modelled: 11
Confidence: 7.9%
Identity: 36%
PDB header:ribosomal protein
Chain: F: PDB Molecule:naf1;
PDBTitle: structure of the gar1 domain of naf1
Phyre2
| 14 |
|
PDB 3ngv chain A
Region: 32 - 58
Aligned: 21
Modelled: 27
Confidence: 7.7%
Identity: 24%
PDB header:transport protein
Chain: A: PDB Molecule:d7 protein;
PDBTitle: crystal structure of anst-d7l1
Phyre2
| 15 |
|
PDB 2pkq chain O domain 2
Region: 13 - 40
Aligned: 28
Modelled: 28
Confidence: 7.4%
Identity: 11%
Fold: FwdE/GAPDH domain-like
Superfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family: GAPDH-like
Phyre2
| 16 |
|
PDB 1rm4 chain A domain 2
Region: 13 - 40
Aligned: 28
Modelled: 28
Confidence: 7.1%
Identity: 0%
Fold: FwdE/GAPDH domain-like
Superfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family: GAPDH-like
Phyre2
| 17 |
|
PDB 1swi chain C
Region: 66 - 78
Aligned: 13
Modelled: 13
Confidence: 5.9%
Identity: 23%
PDB header:leucine zipper
Chain: C: PDB Molecule:gcn4p1;
PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
Phyre2
| 18 |
|
PDB 2w2r chain A
Region: 13 - 20
Aligned: 8
Modelled: 8
Confidence: 5.2%
Identity: 63%
PDB header:viral protein
Chain: A: PDB Molecule:matrix protein;
PDBTitle: structure of the vesicular stomatitis virus matrix protein
Phyre2