| 1 |
|
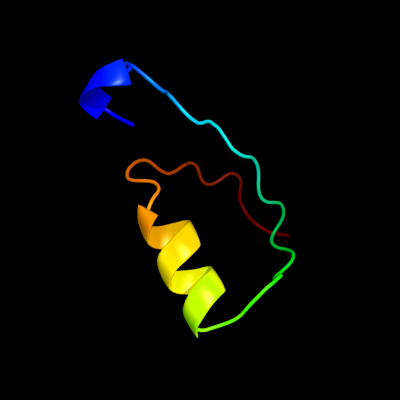
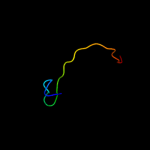
PDB 1e9r chain A
Region: 3 - 40
Aligned: 38
Modelled: 38
Confidence: 50.1%
Identity: 11%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: RecA protein-like (ATPase-domain)
Phyre2
| 2 |
|
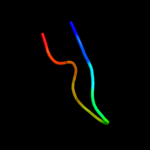
PDB 2pva chain A
Region: 20 - 33
Aligned: 14
Modelled: 14
Confidence: 12.9%
Identity: 36%
Fold: Ntn hydrolase-like
Superfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family: Penicillin V acylase
Phyre2
| 3 |
|
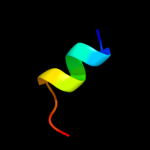
PDB 2hez chain B
Region: 20 - 33
Aligned: 14
Modelled: 14
Confidence: 11.5%
Identity: 21%
PDB header:hydrolase
Chain: B: PDB Molecule:bile salt hydrolase;
PDBTitle: bifidobacterium longum bile salt hydrolase
Phyre2
| 4 |
|
PDB 1shz chain F
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 10.4%
Identity: 13%
PDB header:signaling protein
Chain: F: PDB Molecule:rho guanine nucleotide exchange factor 1;
PDBTitle: crystal structure of the p115rhogef rgrgs domain in a2 complex with galpha(13):galpha(i1) chimera
Phyre2
| 5 |
|
PDB 2bjg chain B
Region: 20 - 33
Aligned: 14
Modelled: 14
Confidence: 10.3%
Identity: 29%
PDB header:hydrolase
Chain: B: PDB Molecule:choloylglycine hydrolase;
PDBTitle: crystal structure of conjugated bile acid hydrolase from2 clostridium perfringens in complex with reaction products3 taurine and deoxycholate
Phyre2
| 6 |
|
PDB 1iap chain A
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 10.3%
Identity: 13%
Fold: Regulator of G-protein signaling, RGS
Superfamily: Regulator of G-protein signaling, RGS
Family: Regulator of G-protein signaling, RGS
Phyre2
| 7 |
|
PDB 1htj chain F
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 9.2%
Identity: 25%
Fold: Regulator of G-protein signaling, RGS
Superfamily: Regulator of G-protein signaling, RGS
Family: Regulator of G-protein signaling, RGS
Phyre2
| 8 |
|
PDB 1htj chain F
Region: 23 - 30
Aligned: 8
Modelled: 8
Confidence: 9.2%
Identity: 25%
PDB header:signaling protein
Chain: F: PDB Molecule:kiaa0380;
PDBTitle: structure of the rgs-like domain from pdz-rhogef
Phyre2
| 9 |
|
PDB 2jxt chain A domain 1
Region: 4 - 28
Aligned: 25
Modelled: 25
Confidence: 9.0%
Identity: 12%
Fold: RplX-like
Superfamily: RplX-like
Family: RplX-like
Phyre2
| 10 |
|
PDB 1tu3 chain J
Region: 17 - 25
Aligned: 9
Modelled: 9
Confidence: 8.7%
Identity: 33%
PDB header:protein transport
Chain: J: PDB Molecule:rab gtpase binding effector protein 1;
PDBTitle: crystal structure of rab5 complex with rabaptin5 c-terminal2 domain
Phyre2
| 11 |
|
PDB 3aqb chain C
Region: 20 - 35
Aligned: 14
Modelled: 16
Confidence: 8.4%
Identity: 50%
PDB header:transferase
Chain: C: PDB Molecule:component a of hexaprenyl diphosphate synthase;
PDBTitle: m. luteus b-p 26 heterodimeric hexaprenyl diphosphate synthase in2 complex with magnesium
Phyre2
| 12 |
|
PDB 2f4i chain A domain 1
Region: 39 - 64
Aligned: 26
Modelled: 26
Confidence: 7.4%
Identity: 15%
Fold: OB-fold
Superfamily: TM0957-like
Family: TM0957-like
Phyre2
| 13 |
|
PDB 3ipj chain B
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 6.6%
Identity: 40%
PDB header:transferase
Chain: B: PDB Molecule:pts system, iiabc component;
PDBTitle: the crystal structure of one domain of the pts system, iiabc component2 from clostridium difficile
Phyre2
| 14 |
|
PDB 2oqc chain B
Region: 22 - 33
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 42%
PDB header:hydrolase
Chain: B: PDB Molecule:penicillin v acylase;
PDBTitle: crystal structure of penicillin v acylase from bacillus subtilis
Phyre2
| 15 |
|
PDB 1iba chain A
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 40%
PDB header:phoshphotransferase
Chain: A: PDB Molecule:glucose permease;
PDBTitle: glucose permease (domain iib), nmr, 11 structures
Phyre2
| 16 |
|
PDB 3hbc chain A
Region: 22 - 33
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 42%
PDB header:hydrolase
Chain: A: PDB Molecule:choloylglycine hydrolase;
PDBTitle: crystal structure of choloylglycine hydrolase from bacteroides2 thetaiotaomicron vpi
Phyre2
| 17 |
|
PDB 3bp8 chain C domain 1
Region: 28 - 37
Aligned: 10
Modelled: 10
Confidence: 5.2%
Identity: 40%
Fold: Homing endonuclease-like
Superfamily: Glucose permease domain IIB
Family: Glucose permease domain IIB
Phyre2