1 c3mkuA_
100.0
55
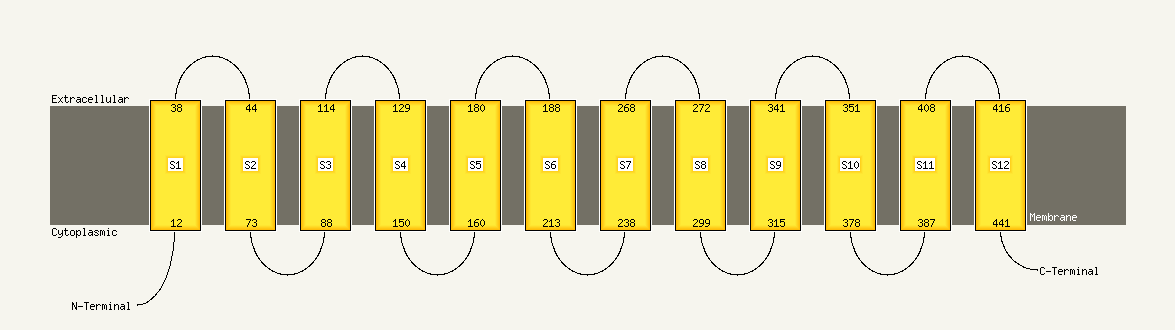
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
2 d1pw4a_
18.8
12
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter3 d2iuba2
10.0
18
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region4 c3g43F_
9.7
8
PDB header: metal binding proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel subunitPDBTitle: crystal structure of the calmodulin-bound cav1.2 c-terminal2 regulatory domain dimer
5 c1wrgA_
9.6
11
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, beta chain;PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
6 d1lghb_
8.8
8
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits7 c3ipdB_
8.7
10
PDB header: exocytosisChain: B: PDB Molecule: syntaxin-1a;PDBTitle: helical extension of the neuronal snare complex into the2 membrane, spacegroup i 21 21 21
8 c3hzqA_
8.3
16
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
9 c2ketA_
7.9
8
PDB header: antibioticChain: A: PDB Molecule: cathelicidin-6;PDBTitle: solution structure of bmap-27
10 d1m56d_
7.7
11
Fold: Single transmembrane helixSuperfamily: Bacterial aa3 type cytochrome c oxidase subunit IVFamily: Bacterial aa3 type cytochrome c oxidase subunit IV11 c1novE_
7.5
15
PDB header: virusChain: E: PDB Molecule: nodamura virus coat proteins;PDBTitle: nodamura virus
12 c2z2qF_
7.4
0
PDB header: virus/rnaChain: F: PDB Molecule: coat protein gamma;PDBTitle: crystal structure of flock house virus
13 d2d5ba1
7.2
8
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases14 d1u61a_
7.1
13
Fold: RNase III domain-likeSuperfamily: RNase III domain-likeFamily: RNase III catalytic domain-like15 d1v54c_
6.9
10
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like16 d1jo5a_
6.7
16
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits17 c2hydB_
6.6
11
PDB header: transport proteinChain: B: PDB Molecule: abc transporter homolog;PDBTitle: multidrug abc transporter sav1866
18 d1w7ab4
6.3
50
Fold: MutS N-terminal domain-likeSuperfamily: DNA repair protein MutS, domain IFamily: DNA repair protein MutS, domain I19 c2l35A_
6.3
7
PDB header: protein bindingChain: A: PDB Molecule: dap12-nkg2c_tm;PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
20 c2gslE_
6.3
11
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: hypothetical protein;PDBTitle: x-ray crystal structure of protein fn1578 from fusobacterium2 nucleatum. northeast structural genomics consortium target nr1.
21 c2k6xA_
not modelled
6.2
20
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: autoregulation of a group 1 bacterial sigma factor involves2 the formation of a region 1.1- induced compacted structure
22 d1pf4a2
not modelled
6.0
13
Fold: ABC transporter transmembrane regionSuperfamily: ABC transporter transmembrane regionFamily: ABC transporter transmembrane region23 c1m46B_
not modelled
5.8
22
PDB header: cell cycle proteinChain: B: PDB Molecule: iq4 motif from myo2p, a class v myosin;PDBTitle: crystal structure of mlc1p bound to iq4 of myo2p, a class v2 myosin
24 c1tiiC_
not modelled
5.7
10
PDB header: enterotoxinChain: C: PDB Molecule: heat labile enterotoxin type iib;PDBTitle: escherichia coli heat labile enterotoxin type iib
25 d1kbhb_
not modelled
5.5
40
Fold: Nuclear receptor coactivator interlocking domainSuperfamily: Nuclear receptor coactivator interlocking domainFamily: Nuclear receptor coactivator interlocking domain26 d1eysh2
not modelled
5.4
7
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region27 d1nvpb_
not modelled
5.4
17
Fold: Transcription factor IIA (TFIIA), alpha-helical domainSuperfamily: Transcription factor IIA (TFIIA), alpha-helical domainFamily: Transcription factor IIA (TFIIA), alpha-helical domain28 c1nvpB_
not modelled
5.4
17
PDB header: transcription/dnaChain: B: PDB Molecule: transcription initiation factor iia alpha chain;PDBTitle: human tfiia/tbp/dna complex
29 c1b9uA_
not modelled
5.4
18
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
30 d1nkzb_
not modelled
5.2
12
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits31 c1unyA_
not modelled
5.2
15
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
32 d1qlec_
not modelled
5.1
13
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like33 d1zoqc1
not modelled
5.1
40
Fold: Nuclear receptor coactivator interlocking domainSuperfamily: Nuclear receptor coactivator interlocking domainFamily: Nuclear receptor coactivator interlocking domain34 c2i7uA_
not modelled
5.1
42
PDB header: de novo protein/ligand binding proteinChain: A: PDB Molecule: four-alpha-helix bundle;PDBTitle: structural and dynamical analysis of a four-alpha-helix2 bundle with designed anesthetic binding pockets
35 d2cqna1
not modelled
5.1
23
Fold: Another 3-helical bundleSuperfamily: FF domainFamily: FF domain36 c1iojA_
not modelled
5.1
43
PDB header: apolipoproteinChain: A: PDB Molecule: apoc-i;PDBTitle: human apolipoprotein c-i, nmr, 18 structures
37 c2khgA_
not modelled
5.0
22
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in tfe
38 c2khfA_
not modelled
5.0
22
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in dpc-micelles