| 1 |
|
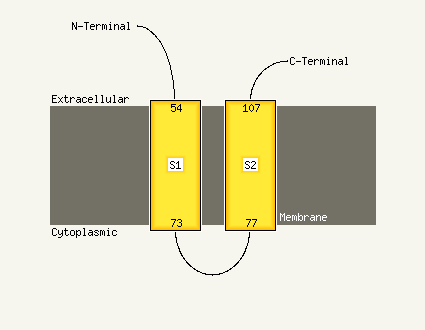
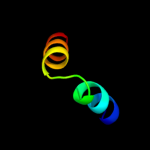
PDB 2gv5 chain C
Region: 2 - 24
Aligned: 23
Modelled: 23
Confidence: 20.5%
Identity: 39%
PDB header:cell cycle
Chain: C: PDB Molecule:sfi1p;
PDBTitle: crystal structure of sfi1p/cdc31p complex
Phyre2
| 2 |
|
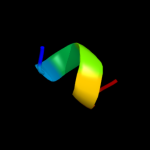
PDB 1eys chain H domain 2
Region: 51 - 67
Aligned: 17
Modelled: 17
Confidence: 10.8%
Identity: 24%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 3 |
|
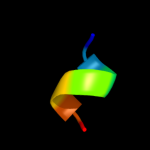
PDB 3pcq chain X
Region: 46 - 62
Aligned: 17
Modelled: 17
Confidence: 8.8%
Identity: 24%
PDB header:photosynthesis
Chain: X: PDB Molecule:photosystem i 4.8k protein;
PDBTitle: femtosecond x-ray protein nanocrystallography
Phyre2
| 4 |
|
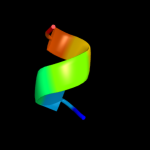
PDB 2w84 chain B
Region: 6 - 19
Aligned: 14
Modelled: 14
Confidence: 8.3%
Identity: 43%
PDB header:protein transport
Chain: B: PDB Molecule:peroxisomal targeting signal 1 receptor;
PDBTitle: structure of pex14 in compex with pex5
Phyre2
| 5 |
|
PDB 1bhb chain A
Region: 48 - 71
Aligned: 24
Modelled: 24
Confidence: 8.3%
Identity: 13%
PDB header:photoreceptor
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
Phyre2
| 6 |
|
PDB 3cxj chain B
Region: 34 - 56
Aligned: 23
Modelled: 23
Confidence: 8.0%
Identity: 26%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein from2 methanothermobacter thermautotrophicus
Phyre2
| 7 |
|
PDB 2fiq chain A domain 1
Region: 24 - 43
Aligned: 20
Modelled: 16
Confidence: 7.9%
Identity: 25%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: GatZ-like
Phyre2
| 8 |
|
PDB 2pco chain A
Region: 70 - 76
Aligned: 7
Modelled: 7
Confidence: 7.7%
Identity: 29%
PDB header:toxin
Chain: A: PDB Molecule:latarcin-1;
PDBTitle: spatial structure and membrane permeabilization for2 latarcin-1, a spider antimicrobial peptide
Phyre2
| 9 |
|
PDB 1vdi chain A
Region: 70 - 76
Aligned: 7
Modelled: 7
Confidence: 6.8%
Identity: 57%
PDB header:contractile protein
Chain: A: PDB Molecule:troponin i, fast skeletal muscle;
PDBTitle: solution structure of actin-binding domain of troponin in2 ca2+-free state
Phyre2
| 10 |
|
PDB 1riq chain A domain 1
Region: 19 - 28
Aligned: 10
Modelled: 10
Confidence: 6.5%
Identity: 60%
Fold: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Superfamily: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Family: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Phyre2
| 11 |
|
PDB 1vf5 chain R
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
PDB header:photosynthesis
Chain: R: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
Phyre2
| 12 |
|
PDB 2e74 chain E domain 1
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
Fold: Single transmembrane helix
Superfamily: PetL subunit of the cytochrome b6f complex
Family: PetL subunit of the cytochrome b6f complex
Phyre2
| 13 |
|
PDB 2e75 chain E
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
Phyre2
| 14 |
|
PDB 2e74 chain E
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
Phyre2
| 15 |
|
PDB 2e76 chain E
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
PDB header:photosynthesis
Chain: E: PDB Molecule:cytochrome b6-f complex subunit 6;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
Phyre2
| 16 |
|
PDB 1vf5 chain E
Region: 56 - 62
Aligned: 7
Modelled: 7
Confidence: 6.3%
Identity: 57%
PDB header:photosynthesis
Chain: E: PDB Molecule:protein pet l;
PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
Phyre2