| 1 |
|
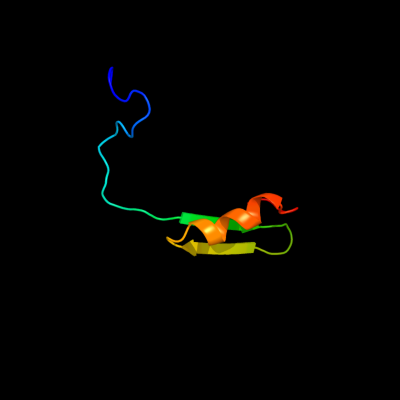
PDB 2iub chain A domain 1
Region: 17 - 65
Aligned: 49
Modelled: 49
Confidence: 35.7%
Identity: 27%
Fold: CorA soluble domain-like
Superfamily: CorA soluble domain-like
Family: CorA soluble domain-like
Phyre2
| 2 |
|
PDB 2bbj chain B
Region: 17 - 65
Aligned: 49
Modelled: 49
Confidence: 22.0%
Identity: 27%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 3 |
|
PDB 1dml chain A domain 1
Region: 14 - 39
Aligned: 24
Modelled: 26
Confidence: 11.5%
Identity: 33%
Fold: DNA clamp
Superfamily: DNA clamp
Family: DNA polymerase processivity factor
Phyre2
| 4 |
|
PDB 3fo8 chain D
Region: 19 - 37
Aligned: 19
Modelled: 19
Confidence: 7.5%
Identity: 37%
PDB header:viral protein
Chain: D: PDB Molecule:tail sheath protein gp18;
PDBTitle: crystal structure of the bacteriophage t4 tail sheath2 protein, protease resistant fragment gp18pr
Phyre2
| 5 |
|
PDB 3nkg chain A
Region: 56 - 71
Aligned: 16
Modelled: 16
Confidence: 7.1%
Identity: 50%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein geba250068378;
PDBTitle: crystal structure of geba250068378 from sulfurospirillum deleyianum
Phyre2
| 6 |
|
PDB 3ghp chain A
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 6.9%
Identity: 75%
PDB header:structural protein
Chain: A: PDB Molecule:cellulosomal scaffoldin adaptor protein b;
PDBTitle: structure of the second type ii cohesin module from the2 adaptor scaa scaffoldin of acetivibrio cellulolyticus3 (including long c-terminal linker)
Phyre2
| 7 |
|
PDB 1t4z chain A
Region: 63 - 72
Aligned: 10
Modelled: 10
Confidence: 6.7%
Identity: 30%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: KaiB-like
Phyre2
| 8 |
|
PDB 2bm3 chain A domain 1
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 6.5%
Identity: 75%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family III
Phyre2
| 9 |
|
PDB 1zv9 chain A domain 1
Region: 28 - 35
Aligned: 8
Modelled: 6
Confidence: 6.5%
Identity: 50%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family III
Phyre2
| 10 |
|
PDB 1tyj chain A domain 1
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 6.2%
Identity: 63%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Carbohydrate-binding domain
Family: Cellulose-binding domain family III
Phyre2
| 11 |
|
PDB 2b59 chain A
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 6.1%
Identity: 75%
PDB header:hydrolase/structural protein
Chain: A: PDB Molecule:cog1196: chromosome segregation atpases;
PDBTitle: the type ii cohesin dockerin complex
Phyre2
| 12 |
|
PDB 3fnk chain A
Region: 28 - 35
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 75%
PDB header:structural protein
Chain: A: PDB Molecule:cellulosomal scaffoldin adaptor protein b;
PDBTitle: crystal structure of the second type ii cohesin module from2 the cellulosomal adaptor scaa scaffoldin of acetivibrio3 cellulolyticus
Phyre2
| 13 |
|
PDB 3izb chain T
Region: 53 - 59
Aligned: 7
Modelled: 7
Confidence: 5.9%
Identity: 71%
PDB header:ribosome
Chain: T: PDB Molecule:40s ribosomal protein rps21 (s21e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 14 |
|
PDB 1dml chain G
Region: 14 - 39
Aligned: 24
Modelled: 26
Confidence: 5.4%
Identity: 33%
PDB header:dna binding protein/transferase
Chain: G: PDB Molecule:dna polymerase processivity factor;
PDBTitle: crystal structure of herpes simplex ul42 bound to the c-2 terminus of hsv pol
Phyre2
| 15 |
|
PDB 3fcg chain B
Region: 16 - 37
Aligned: 22
Modelled: 22
Confidence: 5.4%
Identity: 23%
PDB header:membrane protein, protein transport
Chain: B: PDB Molecule:f1 capsule-anchoring protein;
PDBTitle: crystal structure analysis of the middle domain of the2 caf1a usher
Phyre2