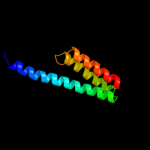
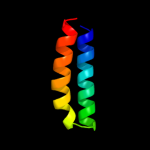
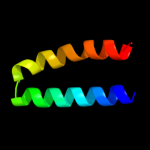
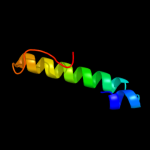
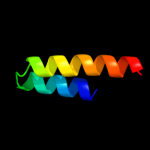
1 d2gy9t1
100.0
100
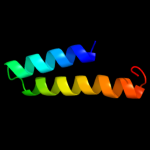
Fold: Spectrin repeat-likeSuperfamily: Ribosomal protein S20Family: Ribosomal protein S202 d2uubt1
100.0
33
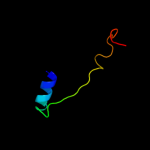
Fold: Spectrin repeat-likeSuperfamily: Ribosomal protein S20Family: Ribosomal protein S203 c3bbnT_
100.0
35
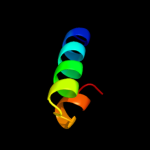
PDB header: ribosomeChain: T: PDB Molecule: ribosomal protein s20;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
4 d1h6qa_
26.7
14
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: Translationally controlled tumor protein TCTP (histamine-releasing factor)5 d1txja_
24.4
9
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: Translationally controlled tumor protein TCTP (histamine-releasing factor)6 c2oxoA_
21.7
23
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: crystallization and structure determination of the core-2 binding domain of bacteriophage lambda integrase
7 c3dzaB_
19.8
15
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized putative membrane protein;PDBTitle: crystal structure of a putative membrane protein of unknown function2 (yfdx) from klebsiella pneumoniae subsp. at 1.65 a resolution
8 c2khvA_
18.9
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of protein nmul_a0922 from2 nitrosospira multiformis. northeast structural genomics3 consortium target nmr38b.
9 c2kj8A_
18.2
16
PDB header: dna binding proteinChain: A: PDB Molecule: putative prophage cps-53 integrase;PDBTitle: nmr structure of fragment 87-196 from the putative phage2 integrase ints of e. coli: northeast structural genomics3 consortium target er652a, psi-2
10 d2k54a1
17.8
23
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Atu0742-like11 d1yz1a1
17.2
7
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: Translationally controlled tumor protein TCTP (histamine-releasing factor)12 c3k1sE_
16.3
18
PDB header: transferaseChain: E: PDB Molecule: pts system, cellobiose-specific iia component;PDBTitle: crystal structure of the pts cellobiose specific enzyme iia2 from bacillus anthracis
13 d2oo2a1
13.6
16
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: AF1782-likeFamily: AF1782-like14 c2dl1A_
13.2
21
PDB header: protein transportChain: A: PDB Molecule: spartin;PDBTitle: solution structure of the mit domain from human spartin
15 c1xb2B_
13.1
28
PDB header: translationChain: B: PDB Molecule: elongation factor ts, mitochondrial;PDBTitle: crystal structure of bos taurus mitochondrial elongation2 factor tu/ts complex
16 d3en8a1
12.9
24
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Rpa4348-like17 c3lysC_
12.9
10
PDB header: recombinationChain: C: PDB Molecule: prophage pi2 protein 01, integrase;PDBTitle: crystal structure of the n-terminal domain of the prophage2 pi2 protein 01 (integrase) from lactococcus lactis,3 northeast structural genomics consortium target kr124f
18 c2kj9A_
10.3
15
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: nmr structure of intb phage-integrase-like protein fragment2 90-199 from erwinia carotova subsp. atroseptica: northeast3 structural genomics consortium target ewr217e
19 d1h7ca_
10.0
8
Fold: Spectrin repeat-likeSuperfamily: Tubulin chaperone cofactor AFamily: Tubulin chaperone cofactor A20 c3soyA_
9.5
13
PDB header: membrane proteinChain: A: PDB Molecule: ntf2-like superfamily protein;PDBTitle: nuclear transport factor 2 (ntf2-like) superfamily protein from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
21 c2khqA_
not modelled
9.1
17
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: solution nmr structure of a phage integrase ssp19472 fragment 59-159 from staphylococcus saprophyticus,3 northeast structural genomics consortium target syr103b
22 c3kspA_
not modelled
7.8
14
PDB header: unknown functionChain: A: PDB Molecule: calcium/calmodulin-dependent kinase ii association domain;PDBTitle: crystal structure of a putative ca/calmodulin-dependent kinase ii2 association domain (exig_1688) from exiguobacterium sibiricum 255-153 at 2.59 a resolution
23 d1ohpa1
not modelled
7.3
28
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ketosteroid isomerase-like24 c3l8rA_
not modelled
6.9
10
PDB header: transferaseChain: A: PDB Molecule: putative pts system, cellobiose-specific iiaPDBTitle: the crystal structure of ptca from s. mutans
25 d2a15a1
not modelled
6.7
12
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ketosteroid isomerase-like26 d1u00a1
not modelled
6.6
15
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Heat shock protein 70kD (HSP70), C-terminal subdomainFamily: Heat shock protein 70kD (HSP70), C-terminal subdomain27 c2odmA_
not modelled
6.6
19
PDB header: unknown functionChain: A: PDB Molecule: upf0358 protein mw0995;PDBTitle: crystal structure of s. aureus ylan, an essential leucine rich protein2 involved in the control of cell shape
28 d2f86b1
not modelled
6.2
10
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Association domain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A29 c1wcrA_
not modelled
6.0
15
PDB header: transferaseChain: A: PDB Molecule: pts system, n, n'-diacetylchitobiose-specificPDBTitle: trimeric structure of the enzyme iia from escherichia coli2 phosphotransferase system specific for n,n'-3 diacetylchitobiose
30 c3kh1B_
not modelled
5.9
15
PDB header: hydrolaseChain: B: PDB Molecule: predicted metal-dependent phosphohydrolase;PDBTitle: crystal structure of predicted metal-dependent2 phosphohydrolase (zp_00055740.2) from magnetospirillum3 magnetotacticum ms-1 at 1.37 a resolution
31 c2kkvA_
not modelled
5.5
18
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: solution nmr structure of an integrase domain from protein2 spa4288 from salmonella enterica, northeast structural3 genomics consortium target slr105h
32 d2ux0a1
not modelled
5.3
23
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Association domain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A33 d1aipc1
not modelled
5.2
29
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TS-N domain34 c3i0yC_
not modelled
5.2
24
PDB header: isomeraseChain: C: PDB Molecule: putative polyketide cyclase;PDBTitle: crystal structure of a putative polyketide cyclase (xcc0381) from2 xanthomonas campestris pv. campestris at 1.50 a resolution