| 1 |
|
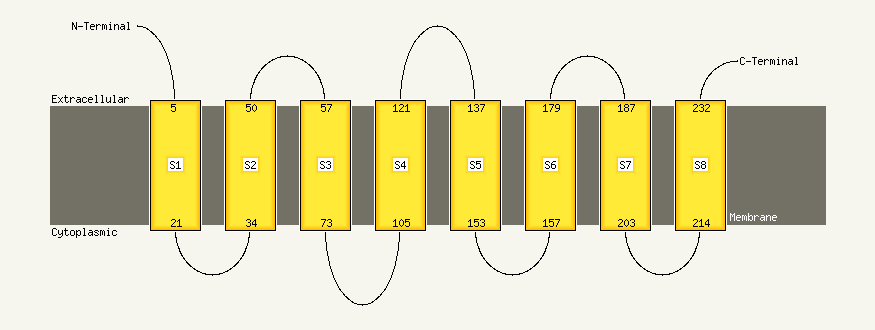
PDB 1kpl chain A
Region: 11 - 92
Aligned: 82
Modelled: 82
Confidence: 12.6%
Identity: 15%
Fold: Clc chloride channel
Superfamily: Clc chloride channel
Family: Clc chloride channel
Phyre2
| 2 |
|
PDB 1rh1 chain A domain 2
Region: 129 - 191
Aligned: 63
Modelled: 63
Confidence: 11.6%
Identity: 14%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 3 |
|
PDB 1dxs chain A
Region: 25 - 38
Aligned: 14
Modelled: 14
Confidence: 11.0%
Identity: 21%
Fold: SAM domain-like
Superfamily: SAM/Pointed domain
Family: SAM (sterile alpha motif) domain
Phyre2
| 4 |
|
PDB 2o34 chain A domain 1
Region: 72 - 126
Aligned: 55
Modelled: 55
Confidence: 10.7%
Identity: 7%
Fold: T-fold
Superfamily: ApbE-like
Family: DVU1097-like
Phyre2
| 5 |
|
PDB 2uwj chain G
Region: 71 - 106
Aligned: 36
Modelled: 36
Confidence: 9.2%
Identity: 3%
PDB header:chaperone
Chain: G: PDB Molecule:type iii export protein pscg;
PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
Phyre2
| 6 |
|
PDB 2ju0 chain B
Region: 215 - 223
Aligned: 9
Modelled: 9
Confidence: 8.4%
Identity: 56%
PDB header:metal binding protein/signaling protein
Chain: B: PDB Molecule:phosphatidylinositol 4-kinase pik1;
PDBTitle: structure of yeast frequenin bound to pdtins 4-kinase
Phyre2
| 7 |
|
PDB 3few chain X
Region: 129 - 191
Aligned: 61
Modelled: 63
Confidence: 7.0%
Identity: 15%
PDB header:immune system
Chain: X: PDB Molecule:colicin s4;
PDBTitle: structure and function of colicin s4, a colicin with a2 duplicated receptor binding domain
Phyre2
| 8 |
|
PDB 1aa7 chain A
Region: 83 - 118
Aligned: 36
Modelled: 36
Confidence: 6.3%
Identity: 11%
Fold: Influenza virus matrix protein M1
Superfamily: Influenza virus matrix protein M1
Family: Influenza virus matrix protein M1
Phyre2
| 9 |
|
PDB 1c7q chain A
Region: 76 - 100
Aligned: 25
Modelled: 25
Confidence: 6.2%
Identity: 20%
Fold: SIS domain
Superfamily: SIS domain
Family: Phosphoglucose isomerase, PGI
Phyre2