| 1 |
|
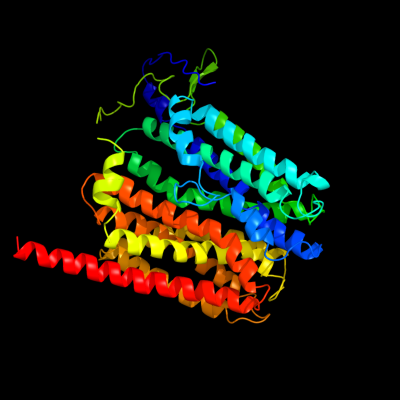
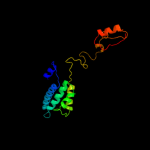
PDB 1pw4 chain A
Region: 5 - 451
Aligned: 424
Modelled: 435
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
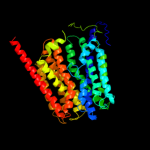
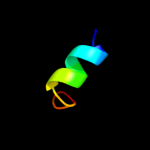
PDB 3o7p chain A
Region: 25 - 441
Aligned: 400
Modelled: 417
Confidence: 100.0%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 3 |
|
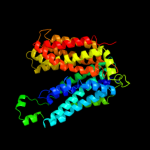
PDB 1pv7 chain A
Region: 20 - 452
Aligned: 411
Modelled: 417
Confidence: 100.0%
Identity: 13%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
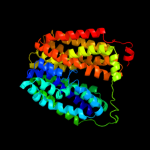
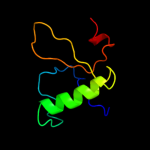
PDB 2gfp chain A
Region: 27 - 438
Aligned: 374
Modelled: 399
Confidence: 100.0%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 27 - 437
Aligned: 401
Modelled: 411
Confidence: 99.9%
Identity: 9%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
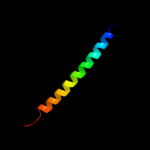
PDB 1r48 chain A
Region: 468 - 497
Aligned: 30
Modelled: 30
Confidence: 97.3%
Identity: 100%
PDB header:transport protein
Chain: A: PDB Molecule:proline/betaine transporter;
PDBTitle: solution structure of the c-terminal cytoplasmic domain2 residues 468-497 of escherichia coli protein prop
Phyre2
| 7 |
|
PDB 3b9y chain A
Region: 322 - 487
Aligned: 160
Modelled: 166
Confidence: 50.5%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 8 |
|
PDB 3org chain B
Region: 339 - 500
Aligned: 160
Modelled: 162
Confidence: 24.5%
Identity: 11%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 9 |
|
PDB 2g9p chain A
Region: 85 - 98
Aligned: 14
Modelled: 14
Confidence: 17.1%
Identity: 50%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 10 |
|
PDB 2l5g chain B
Region: 459 - 496
Aligned: 38
Modelled: 38
Confidence: 8.6%
Identity: 26%
PDB header:transcription regulator
Chain: B: PDB Molecule:putative uncharacterized protein ncor2;
PDBTitle: co-ordinates and 1h, 13c and 15n chemical shift assignments for the2 complex of gps2 53-90 and smrt 167-207
Phyre2
| 11 |
|
PDB 3bz6 chain A domain 2
Region: 430 - 500
Aligned: 69
Modelled: 71
Confidence: 7.5%
Identity: 9%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: PSPTO2686-like
Phyre2
| 12 |
|
PDB 3qnq chain D
Region: 404 - 456
Aligned: 53
Modelled: 53
Confidence: 7.3%
Identity: 15%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 13 |
|
PDB 1a92 chain B
Region: 471 - 496
Aligned: 26
Modelled: 26
Confidence: 6.4%
Identity: 19%
PDB header:leucine zipper
Chain: B: PDB Molecule:delta antigen;
PDBTitle: oligomerization domain of hepatitis delta antigen
Phyre2