| 1 |
|
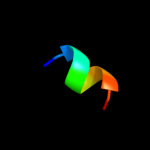
PDB 2r2v chain B
Region: 66 - 84
Aligned: 19
Modelled: 19
Confidence: 19.8%
Identity: 42%
PDB header:de novo protein
Chain: B: PDB Molecule:gcn4 leucine zipper;
PDBTitle: sequence determinants of the topology of the lac repressor2 tetrameric coiled coil
Phyre2
| 2 |
|
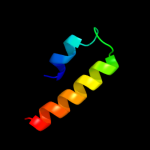
PDB 2laf chain A
Region: 56 - 75
Aligned: 20
Modelled: 20
Confidence: 12.1%
Identity: 30%
PDB header:membrane protein
Chain: A: PDB Molecule:lipoprotein 34;
PDBTitle: nmr solution structure of the n-terminal domain of the e. coli2 lipoprotein bamc
Phyre2
| 3 |
|
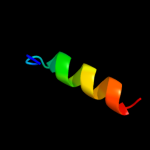
PDB 2k7w chain B
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 11.5%
Identity: 38%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: bax activation is initiated at a novel interaction site
Phyre2
| 4 |
|
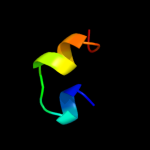
PDB 3kj1 chain B
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 11.3%
Identity: 38%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: mcl-1 in complex with bim bh3 mutant i2da
Phyre2
| 5 |
|
PDB 3kj2 chain B
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 10.9%
Identity: 38%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: mcl-1 in complex with bim bh3 mutant f4ae
Phyre2
| 6 |
|
PDB 3kz0 chain D
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 38%
PDB header:apoptosis
Chain: D: PDB Molecule:mcl-1 specific peptide mb7;
PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
Phyre2
| 7 |
|
PDB 3kz0 chain C
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 9.1%
Identity: 38%
PDB header:apoptosis
Chain: C: PDB Molecule:mcl-1 specific peptide mb7;
PDBTitle: mcl-1 complex with mcl-1-specific selected peptide
Phyre2
| 8 |
|
PDB 1pq1 chain B
Region: 64 - 82
Aligned: 19
Modelled: 19
Confidence: 8.7%
Identity: 32%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl2-like protein 11;
PDBTitle: crystal structure of bcl-xl/bim
Phyre2
| 9 |
|
PDB 3io9 chain B
Region: 64 - 71
Aligned: 8
Modelled: 8
Confidence: 6.3%
Identity: 38%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-like protein 11;
PDBTitle: biml12y in complex with mcl-1
Phyre2
| 10 |
|
PDB 2z15 chain A domain 1
Region: 49 - 81
Aligned: 33
Modelled: 33
Confidence: 6.2%
Identity: 18%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 11 |
|
PDB 3cf4 chain A
Region: 30 - 47
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 22%
PDB header:oxidoreductase
Chain: A: PDB Molecule:acetyl-coa decarboxylase/synthase alpha subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
Phyre2
| 12 |
|
PDB 1ery chain A
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 5.2%
Identity: 31%
Fold: Protozoan pheromone-like
Superfamily: Protozoan pheromone proteins
Family: Protozoan pheromone proteins
Phyre2