| 1 |
|
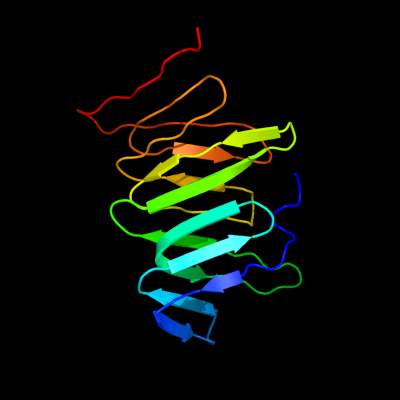
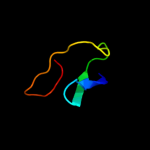
PDB 2r1a chain D
Region: 28 - 166
Aligned: 136
Modelled: 139
Confidence: 100.0%
Identity: 99%
PDB header:transport protein
Chain: D: PDB Molecule:protein yhbn;
PDBTitle: crystal structure of the periplasmic lipopolysaccharide transport2 protein lpta (yhbn), trigonal form
Phyre2
| 2 |
|
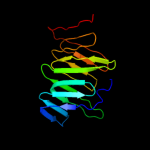
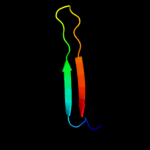
PDB 3my2 chain A
Region: 37 - 165
Aligned: 118
Modelled: 129
Confidence: 98.8%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:lipopolysaccharide export system protein lptc;
PDBTitle: crystal structure of lptc
Phyre2
| 3 |
|
PDB 1g7s chain A domain 1
Region: 145 - 174
Aligned: 30
Modelled: 30
Confidence: 25.0%
Identity: 17%
Fold: Reductase/isomerase/elongation factor common domain
Superfamily: Translation proteins
Family: Elongation factors
Phyre2
| 4 |
|
PDB 1ueb chain A domain 3
Region: 118 - 151
Aligned: 34
Modelled: 34
Confidence: 8.6%
Identity: 15%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 5 |
|
PDB 1xct chain L
Region: 29 - 56
Aligned: 28
Modelled: 28
Confidence: 7.2%
Identity: 14%
PDB header:immune system
Chain: L: PDB Molecule:protein l;
PDBTitle: complex hcv core-fab 19d9d6-protein l mutant (d55a, l57h, y64w) in2 space group p21212
Phyre2
| 6 |
|
PDB 2ptl chain A
Region: 31 - 56
Aligned: 26
Modelled: 26
Confidence: 7.1%
Identity: 15%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Immunoglobulin-binding domains
Family: Immunoglobulin-binding domains
Phyre2
| 7 |
|
PDB 2jmb chain A
Region: 111 - 148
Aligned: 38
Modelled: 38
Confidence: 6.8%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein atu4866;
PDBTitle: solution structure of the protein atu4866 from agrobacterium2 tumefaciens
Phyre2