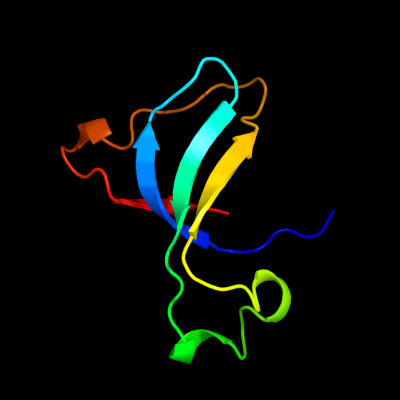
1 c2l55A_
39.5
16
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
2 d1gpra_
29.7
16
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like3 d2f3ga_
28.6
18
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like4 d2gpra_
27.5
18
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like5 c2dchX_
26.8
44
PDB header: hydrolaseChain: X: PDB Molecule: putative homing endonuclease;PDBTitle: crystal structure of archaeal intron-encoded homing endonuclease i-2 tsp061i
6 d1glaf_
24.6
18
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like7 d1apja_
23.8
26
Fold: TB module/8-cys domainSuperfamily: TB module/8-cys domainFamily: TB module/8-cys domain8 c2xgyA_
19.0
17
PDB header: viral protein/isomeraseChain: A: PDB Molecule: relik capsid n-terminal domain;PDBTitle: complex of rabbit endogenous lentivirus (relik)capsid with2 cyclophilin a
9 d1ksqa_
17.7
22
Fold: TB module/8-cys domainSuperfamily: TB module/8-cys domainFamily: TB module/8-cys domain10 c1zeqX_
13.6
3
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
11 c2j5dA_
12.4
39
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles
12 c2d1kC_
11.9
36
PDB header: structural proteinChain: C: PDB Molecule: metastasis suppressor protein 1;PDBTitle: ternary complex of the wh2 domain of mim with actin-dnase i
13 d2cbra_
11.1
27
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like14 d1gl4a1
10.6
13
Fold: GFP-likeSuperfamily: GFP-likeFamily: Domain G2 of nidogen-115 d1fasa_
10.0
57
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins16 d1u0ma2
9.9
13
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like17 d1ve2a1
9.8
41
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase18 c3h41A_
9.3
20
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
19 c1wl5A_
9.0
20
PDB header: transferaseChain: A: PDB Molecule: acetyl-coenzyme a acetyltransferase 2;PDBTitle: human cytosolic acetoacetyl-coa thiolase
20 c2e4mC_
8.7
20
PDB header: toxinChain: C: PDB Molecule: ha-17;PDBTitle: crystal structure of hemagglutinin subcomponent complex (ha-2 33/ha-17) from clostridium botulinum serotype d strain 4947
21 c2e0kA_
not modelled
8.5
35
PDB header: transferaseChain: A: PDB Molecule: precorrin-2 c20-methyltransferase;PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
22 c3nutC_
not modelled
8.2
41
PDB header: transferaseChain: C: PDB Molecule: precorrin-3 methylase;PDBTitle: crystal structure of the methyltransferase cobj
23 d1twda_
not modelled
8.1
100
Fold: TIM beta/alpha-barrelSuperfamily: CutC-likeFamily: CutC-like24 c1h4uA_
not modelled
8.1
13
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
25 c3iwpK_
not modelled
7.6
80
PDB header: metal binding proteinChain: K: PDB Molecule: copper homeostasis protein cutc homolog;PDBTitle: crystal structure of human copper homeostasis protein cutc
26 c2bb3B_
not modelled
7.4
22
PDB header: transferaseChain: B: PDB Molecule: cobalamin biosynthesis precorrin-6y methylase (cbie);PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
27 d1drsa_
not modelled
7.0
38
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Dendroaspin28 d1cnza_
not modelled
6.9
29
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases29 c2kzqA_
not modelled
6.8
25
PDB header: membrane proteinChain: A: PDB Molecule: envelope glycoprotein e2 peptide;PDBTitle: s34r structure
30 c2vu2D_
not modelled
6.7
16
PDB header: transferaseChain: D: PDB Molecule: acetyl-coa acetyltransferase;PDBTitle: biosynthetic thiolase from z. ramigera. complex with s-2 pantetheine-11-pivalate.
31 d1va0a1
not modelled
6.6
41
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase32 d1wyza1
not modelled
6.6
18
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase33 d2eiaa2
not modelled
6.6
16
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain34 d1ycna_
not modelled
6.4
11
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin35 c1cbfA_
not modelled
6.3
35
PDB header: methyltransferaseChain: A: PDB Molecule: cobalt-precorrin-4 transmethylase;PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
36 d1cbfa_
not modelled
6.3
35
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase37 d1niya_
not modelled
6.3
58
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins38 d1mb6a_
not modelled
6.3
33
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins39 c2npnA_
not modelled
6.3
35
PDB header: transferaseChain: A: PDB Molecule: putative cobalamin synthesis related protein;PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
40 c3ndcB_
not modelled
6.2
41
PDB header: transferaseChain: B: PDB Molecule: precorrin-4 c(11)-methyltransferase;PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
41 d1cm7a_
not modelled
6.1
29
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases42 d2bb3a1
not modelled
6.0
25
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase43 c2avuF_
not modelled
6.0
18
PDB header: transcription activatorChain: F: PDB Molecule: flagellar transcriptional activator flhc;PDBTitle: structure of the escherichia coli flhdc complex, a2 prokaryotic heteromeric regulator of transcription
44 d1pkla1
not modelled
5.9
6
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain45 d1v53a1
not modelled
5.8
29
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases46 d1dq3a3
not modelled
5.8
31
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease47 c4a1cX_
not modelled
5.6
27
PDB header: ribosomeChain: X: PDB Molecule: 60s ribosomal protein l32;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
48 d1pjqa2
not modelled
5.6
41
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase49 d1q90g_
not modelled
5.5
44
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex50 c1q90G_
not modelled
5.5
44
PDB header: photosynthesisChain: G: PDB Molecule: cytochrome b6f complex subunit petg;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
51 d2d0oa1
not modelled
5.4
80
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Swiveling domain of dehydratase reactivase alpha subunitFamily: Swiveling domain of dehydratase reactivase alpha subunit52 d1vhva_
not modelled
5.3
12
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase53 c3u1hA_
not modelled
5.2
29
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-isopropylmalate dehydrogenase;PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
54 c2zvbA_
not modelled
5.2
47
PDB header: transferaseChain: A: PDB Molecule: precorrin-3 c17-methyltransferase;PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
55 d1y7ma1
not modelled
5.1
8
Fold: L,D-transpeptidase catalytic domain-likeSuperfamily: L,D-transpeptidase catalytic domain-likeFamily: L,D-transpeptidase catalytic domain-like56 d1ep3b2
not modelled
5.1
40
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domainSuperfamily: Ferredoxin reductase-like, C-terminal NADP-linked domainFamily: Dihydroorotate dehydrogenase B, PyrK subunit57 c3kwpA_
not modelled
5.0
13
PDB header: transferaseChain: A: PDB Molecule: predicted methyltransferase;PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis