| 1 |
|
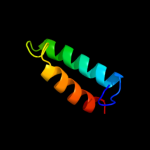
PDB 2hg5 chain D
Region: 182 - 225
Aligned: 42
Modelled: 44
Confidence: 22.5%
Identity: 14%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 2 |
|
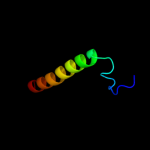
PDB 1jb0 chain L
Region: 178 - 224
Aligned: 45
Modelled: 47
Confidence: 20.1%
Identity: 27%
Fold: Photosystem I reaction center subunit XI, PsaL
Superfamily: Photosystem I reaction center subunit XI, PsaL
Family: Photosystem I reaction center subunit XI, PsaL
Phyre2
| 3 |
|
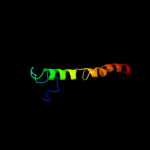
PDB 2i53 chain A domain 1
Region: 268 - 311
Aligned: 44
Modelled: 44
Confidence: 14.4%
Identity: 16%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 4 |
|
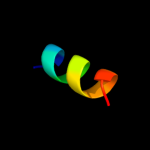
PDB 1f6g chain A
Region: 170 - 225
Aligned: 54
Modelled: 56
Confidence: 12.8%
Identity: 15%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 5 |
|
PDB 1s6x chain A
Region: 223 - 229
Aligned: 7
Modelled: 7
Confidence: 12.2%
Identity: 43%
PDB header:toxin
Chain: A: PDB Molecule:kvap channel;
PDBTitle: solution structure of vstx
Phyre2
| 6 |
|
PDB 1v54 chain J
Region: 142 - 178
Aligned: 37
Modelled: 37
Confidence: 11.6%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIa
Family: Mitochondrial cytochrome c oxidase subunit VIIa
Phyre2
| 7 |
|
PDB 3oa1 chain B
Region: 204 - 221
Aligned: 18
Modelled: 18
Confidence: 10.6%
Identity: 39%
PDB header:chaperone
Chain: B: PDB Molecule:phosphoprotein;
PDBTitle: crystal structure of phosphoprotein/protein p/protein m1 residues 69-2 297 from rabies virus reveals degradation to c-terminal domain only
Phyre2
| 8 |
|
PDB 1flc chain A domain 1
Region: 125 - 138
Aligned: 14
Modelled: 14
Confidence: 10.4%
Identity: 43%
Fold: Viral protein domain
Superfamily: Viral protein domain
Family: Hemagglutinin domain of haemagglutinin-esterase-fusion glycoprotein HEF1
Phyre2
| 9 |
|
PDB 2bs2 chain C domain 1
Region: 219 - 246
Aligned: 28
Modelled: 28
Confidence: 10.3%
Identity: 11%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Fumarate reductase respiratory complex cytochrome b subunit, FrdC
Phyre2
| 10 |
|
PDB 1v74 chain B
Region: 281 - 319
Aligned: 37
Modelled: 39
Confidence: 9.9%
Identity: 22%
Fold: Four-helical up-and-down bundle
Superfamily: Colicin D immunity protein
Family: Colicin D immunity protein
Phyre2
| 11 |
|
PDB 1vyi chain A
Region: 204 - 221
Aligned: 18
Modelled: 18
Confidence: 9.4%
Identity: 39%
Fold: Phosphoprotein M1, C-terminal domain
Superfamily: Phosphoprotein M1, C-terminal domain
Family: Phosphoprotein M1, C-terminal domain
Phyre2
| 12 |
|
PDB 2ivx chain A domain 1
Region: 266 - 311
Aligned: 46
Modelled: 46
Confidence: 9.4%
Identity: 15%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 13 |
|
PDB 2y69 chain W
Region: 142 - 178
Aligned: 37
Modelled: 37
Confidence: 9.1%
Identity: 30%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase polypeptide 7a1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 14 |
|
PDB 2kb1 chain A
Region: 170 - 225
Aligned: 54
Modelled: 56
Confidence: 8.9%
Identity: 17%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 15 |
|
PDB 3kfo chain A
Region: 272 - 283
Aligned: 12
Modelled: 12
Confidence: 8.8%
Identity: 67%
PDB header:protein transport
Chain: A: PDB Molecule:nucleoporin nup133;
PDBTitle: crystal structure of the c-terminal domain from the nuclear pore2 complex component nup133 from saccharomyces cerevisiae
Phyre2
| 16 |
|
PDB 1qhb chain A
Region: 181 - 199
Aligned: 19
Modelled: 19
Confidence: 8.7%
Identity: 21%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 17 |
|
PDB 1up8 chain A
Region: 181 - 199
Aligned: 19
Modelled: 19
Confidence: 8.5%
Identity: 21%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 18 |
|
PDB 1qi9 chain A
Region: 181 - 199
Aligned: 19
Modelled: 19
Confidence: 8.5%
Identity: 21%
Fold: Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily: Acid phosphatase/Vanadium-dependent haloperoxidase
Family: Haloperoxidase (bromoperoxidase)
Phyre2
| 19 |
|
PDB 1iye chain A
Region: 210 - 230
Aligned: 21
Modelled: 21
Confidence: 7.7%
Identity: 29%
Fold: D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily: D-aminoacid aminotransferase-like PLP-dependent enzymes
Family: D-aminoacid aminotransferase-like PLP-dependent enzymes
Phyre2
| 20 |
|
PDB 2hkj chain A domain 2
Region: 288 - 294
Aligned: 7
Modelled: 7
Confidence: 7.4%
Identity: 71%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|