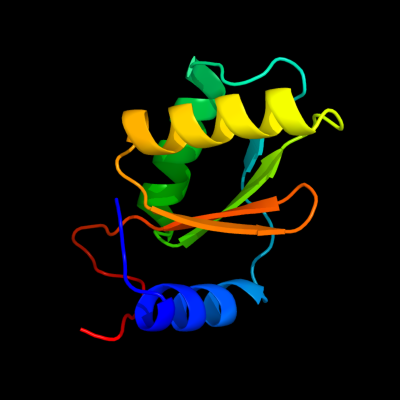
| 1 | d1ln4a_
|
|
 |
100.0 |
98 |
Fold:IF3-like
Superfamily:YhbY-like
Family:YhbY-like |
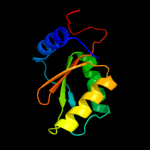
| 2 | d1jo0a_
|
|
 |
100.0 |
75 |
Fold:IF3-like
Superfamily:YhbY-like
Family:YhbY-like |
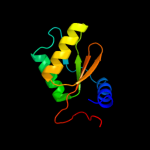
| 3 | d1rq8a_
|
|
 |
100.0 |
38 |
Fold:IF3-like
Superfamily:YhbY-like
Family:YhbY-like |
| 4 | c2e6xD_
|
|
 |
27.4 |
18 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ttha1281;
PDBTitle: x-ray structure of tt1592 from thermus thermophilus hb8
|
| 5 | d2pc6a1
|
|
 |
25.8 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 6 | d1stza1
|
|
 |
16.7 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Heat-inducible transcription repressor HrcA, N-terminal domain |
| 7 | d1iufa1
|
|
 |
15.7 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Centromere-binding |
| 8 | c2pc6C_
|
|
 |
15.6 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 9 | d1nvpb_
|
|
 |
13.9 |
35 |
Fold:Transcription factor IIA (TFIIA), alpha-helical domain
Superfamily:Transcription factor IIA (TFIIA), alpha-helical domain
Family:Transcription factor IIA (TFIIA), alpha-helical domain |
| 10 | c1nvpB_
|
|
 |
13.9 |
35 |
PDB header:transcription/dna
Chain: B: PDB Molecule:transcription initiation factor iia alpha chain;
PDBTitle: human tfiia/tbp/dna complex
|
| 11 | d1wmub_
|
|
 |
13.1 |
15 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 12 | c1lc3A_
|
|
 |
13.1 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biliverdin reductase a;
PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
|
| 13 | d1nh2b_
|
|
 |
12.3 |
18 |
Fold:Transcription factor IIA (TFIIA), alpha-helical domain
Superfamily:Transcription factor IIA (TFIIA), alpha-helical domain
Family:Transcription factor IIA (TFIIA), alpha-helical domain |
| 14 | d1s16a2
|
|
 |
11.7 |
10 |
Fold:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Superfamily:ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase
Family:DNA gyrase/MutL, N-terminal domain |
| 15 | d1nh9a_
|
|
 |
11.5 |
17 |
Fold:IF3-like
Superfamily:AlbA-like
Family:DNA-binding protein AlbA |
| 16 | c1zzgB_
|
|
 |
11.5 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:glucose-6-phosphate isomerase;
PDBTitle: crystal structure of hypothetical protein tt0462 from thermus2 thermophilus hb8
|
| 17 | d2f1fa2
|
|
 |
11.3 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 18 | d1fhjb_
|
|
 |
10.7 |
23 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 19 | d2fgca1
|
|
 |
10.6 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 20 | d1cg5b_
|
|
 |
10.6 |
38 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 21 | d1spgb_ |
|
not modelled |
10.5 |
8 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 22 | c3ka5A_ |
|
not modelled |
10.2 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:ribosome-associated protein y (psrp-1);
PDBTitle: crystal structure of ribosome-associated protein y (psrp-1)2 from clostridium acetobutylicum. northeast structural3 genomics consortium target id car123a
|
| 23 | c3rbvA_ |
|
not modelled |
9.2 |
10 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 24 | c3e9mC_ |
|
not modelled |
8.6 |
8 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 25 | d1l3la1 |
|
not modelled |
8.3 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 26 | d1oela2 |
|
not modelled |
8.0 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 27 | d3bj1b1 |
|
not modelled |
7.8 |
0 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 28 | c2yu3A_ |
|
not modelled |
7.4 |
30 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase iii 39 kda
PDBTitle: solution structure of the domain swapped wingedhelix in dna-2 directed rna polymerase iii 39 kda polypeptide
|
| 29 | d2d5xb1 |
|
not modelled |
7.3 |
23 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 30 | d1srva_ |
|
not modelled |
7.1 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 31 | c2fgcA_ |
|
not modelled |
7.1 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
| 32 | d1lc0a1 |
|
not modelled |
7.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 33 | d1we3a2 |
|
not modelled |
6.8 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 34 | c2rrnA_ |
|
not modelled |
6.8 |
16 |
PDB header:protein transport
Chain: A: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: solution structure of secdf periplasmic domain p4
|
| 35 | d1dk7a_ |
|
not modelled |
6.5 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 36 | c3k2tA_ |
|
not modelled |
6.5 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo2511 protein;
PDBTitle: crystal structure of lmo2511 protein from listeria2 monocytogenes, northeast structural genomics consortium3 target lkr84a
|
| 37 | c3mvcB_ |
|
not modelled |
6.5 |
15 |
PDB header:electron transport
Chain: B: PDB Molecule:globin protein 6;
PDBTitle: high resolution crystal structure of the heme domain of glb-6 from c.2 elegans
|
| 38 | c1tltB_ |
|
not modelled |
6.4 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase (virulence factor mvim homolog);
PDBTitle: crystal structure of a putative oxidoreductase (virulence factor mvim2 homolog)
|
| 39 | c2nuhA_ |
|
not modelled |
6.4 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:periplasmic divalent cation tolerance protein;
PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
|
| 40 | c2h9uA_ |
|
not modelled |
6.4 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna/rna-binding protein alba 2;
PDBTitle: crystal structure of the archaea specific dna binding protein
|
| 41 | d1kida_ |
|
not modelled |
6.3 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 42 | d2d0ob1 |
|
not modelled |
6.2 |
13 |
Fold:Anticodon-binding domain-like
Superfamily:B12-dependent dehydatase associated subunit
Family:Dehydratase-reactivating factor beta subunit |
| 43 | d1nfja_ |
|
not modelled |
6.1 |
19 |
Fold:IF3-like
Superfamily:AlbA-like
Family:DNA-binding protein AlbA |
| 44 | d1sjpa2 |
|
not modelled |
6.0 |
12 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 45 | d1fsla_ |
|
not modelled |
6.0 |
15 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 46 | d1tw4a_ |
|
not modelled |
5.8 |
32 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 47 | c3em0A_ |
|
not modelled |
5.8 |
21 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:ileal bile acid-binding protein;
PDBTitle: crystal structure of zebrafish ileal bile acid-bindin protein2 complexed with cholic acid (crystal form b).
|
| 48 | d1r6bx3 |
|
not modelled |
5.7 |
23 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 49 | d1ofha_ |
|
not modelled |
5.6 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 50 | d1ydha_ |
|
not modelled |
5.6 |
21 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 51 | d2isba1 |
|
not modelled |
5.5 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:FumA C-terminal domain-like
Family:FumA C-terminal domain-like |
| 52 | d1bxni_ |
|
not modelled |
5.4 |
27 |
Fold:RuBisCO, small subunit
Superfamily:RuBisCO, small subunit
Family:RuBisCO, small subunit |
| 53 | d1jg5a_ |
|
not modelled |
5.4 |
22 |
Fold:GTP cyclohydrolase I feedback regulatory protein, GFRP
Superfamily:GTP cyclohydrolase I feedback regulatory protein, GFRP
Family:GTP cyclohydrolase I feedback regulatory protein, GFRP |
| 54 | d2nrka1 |
|
not modelled |
5.4 |
25 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:GrpB-like |
| 55 | d1owla2 |
|
not modelled |
5.4 |
17 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 56 | d1np7a2 |
|
not modelled |
5.3 |
17 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 57 | d1ub3a_ |
|
not modelled |
5.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 58 | d2cvea1 |
|
not modelled |
5.3 |
25 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
| 59 | d2a4aa1 |
|
not modelled |
5.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 60 | d1a04a1 |
|
not modelled |
5.2 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
| 61 | c1tezB_ |
|
not modelled |
5.2 |
17 |
PDB header:lyase/dna
Chain: B: PDB Molecule:deoxyribodipyrimidine photolyase;
PDBTitle: complex between dna and the dna photolyase from anacystis nidulans
|