| 1 |
|
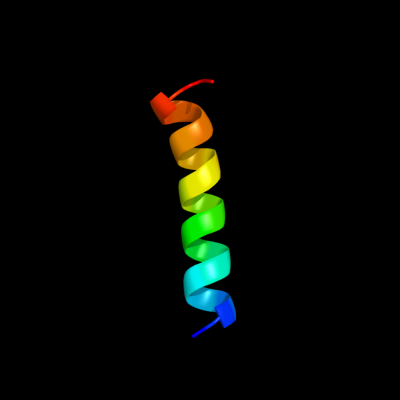
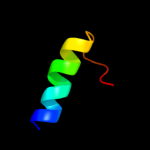
PDB 1vry chain A
Region: 53 - 75
Aligned: 23
Modelled: 23
Confidence: 37.3%
Identity: 35%
PDB header:membrane protein
Chain: A: PDB Molecule:glycine receptor alpha-1 chain;
PDBTitle: second and third transmembrane domains of the alpha-12 subunit of human glycine receptor
Phyre2
| 2 |
|
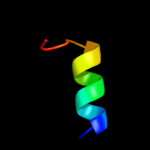
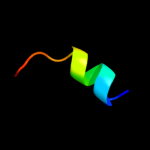
PDB 1lgh chain B
Region: 64 - 78
Aligned: 15
Modelled: 15
Confidence: 29.6%
Identity: 40%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 3 |
|
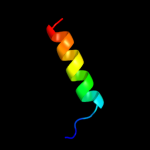
PDB 1eys chain H
Region: 43 - 62
Aligned: 20
Modelled: 20
Confidence: 19.7%
Identity: 35%
PDB header:electron transport
Chain: H: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
Phyre2
| 4 |
|
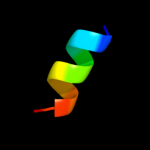
PDB 1ho8 chain A
Region: 50 - 60
Aligned: 11
Modelled: 11
Confidence: 19.4%
Identity: 36%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: Regulatory subunit H of the V-type ATPase
Phyre2
| 5 |
|
PDB 1jo5 chain A
Region: 64 - 78
Aligned: 15
Modelled: 15
Confidence: 19.3%
Identity: 47%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
| 6 |
|
PDB 1eys chain H domain 2
Region: 43 - 68
Aligned: 24
Modelled: 26
Confidence: 14.3%
Identity: 29%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 7 |
|
PDB 1ppj chain C domain 1
Region: 47 - 72
Aligned: 26
Modelled: 26
Confidence: 13.7%
Identity: 42%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 8 |
|
PDB 1m98 chain A domain 1
Region: 35 - 54
Aligned: 20
Modelled: 20
Confidence: 10.0%
Identity: 25%
Fold: Orange carotenoid protein, N-terminal domain
Superfamily: Orange carotenoid protein, N-terminal domain
Family: Orange carotenoid protein, N-terminal domain
Phyre2
| 9 |
|
PDB 3d1d chain C
Region: 43 - 55
Aligned: 13
Modelled: 13
Confidence: 8.7%
Identity: 46%
PDB header:nuclear protein
Chain: C: PDB Molecule:rna-induced transcriptional silencing complex
PDBTitle: hexagonal crystal structure of tas3 c-terminal alpha motif
Phyre2
| 10 |
|
PDB 2ww9 chain B
Region: 59 - 69
Aligned: 11
Modelled: 11
Confidence: 7.9%
Identity: 36%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sss1;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2
| 11 |
|
PDB 1wrg chain A
Region: 57 - 78
Aligned: 22
Modelled: 22
Confidence: 7.7%
Identity: 27%
PDB header:membrane protein
Chain: A: PDB Molecule:light-harvesting protein b-880, beta chain;
PDBTitle: light-harvesting complex 1 beta subunit from wild-type2 rhodospirillum rubrum
Phyre2
| 12 |
|
PDB 2knc chain A
Region: 12 - 24
Aligned: 13
Modelled: 13
Confidence: 7.2%
Identity: 31%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 13 |
|
PDB 1fqv chain A domain 1
Region: 15 - 26
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 58%
Fold: F-box domain
Superfamily: F-box domain
Family: F-box domain
Phyre2