| 1 |
|
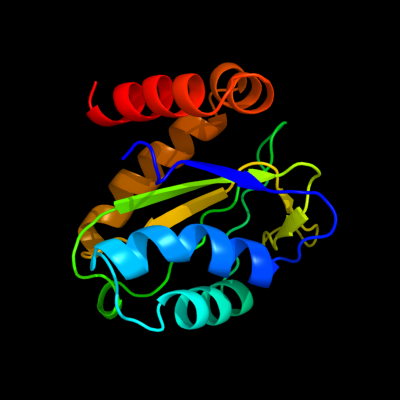
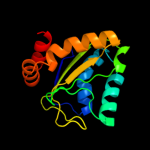
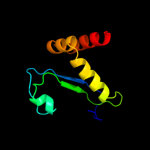
PDB 1xiz chain A
Region: 2 - 147
Aligned: 146
Modelled: 146
Confidence: 100.0%
Identity: 21%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 2 |
|
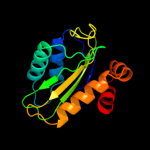
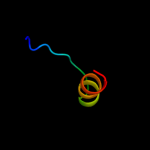
PDB 3oxp chain B
Region: 1 - 144
Aligned: 140
Modelled: 144
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 3 |
|
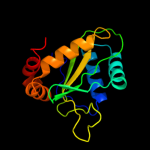
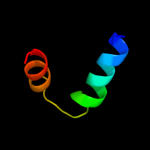
PDB 3oxp chain A
Region: 1 - 144
Aligned: 140
Modelled: 144
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 4 |
|
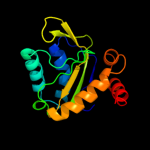
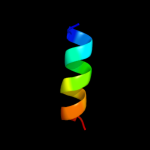
PDB 2oq3 chain A
Region: 1 - 143
Aligned: 138
Modelled: 139
Confidence: 100.0%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific cryptic phosphotransferase
PDBTitle: solution structure of the mannitol- specific cryptic2 phosphotransferase enzyme iia cmtb from escherichia coli
Phyre2
| 5 |
|
PDB 3urr chain B
Region: 1 - 144
Aligned: 139
Modelled: 141
Confidence: 100.0%
Identity: 24%
PDB header:transferase
Chain: B: PDB Molecule:pts iia-like nitrogen-regulatory protein ptsn;
PDBTitle: structure of pts iia-like nitrogen-regulatory protein ptsn (bth_i0484)2 (ptsn)
Phyre2
| 6 |
|
PDB 1a3a chain A
Region: 1 - 149
Aligned: 139
Modelled: 149
Confidence: 100.0%
Identity: 22%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 7 |
|
PDB 3bjv chain A
Region: 1 - 148
Aligned: 142
Modelled: 148
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:rmpa;
PDBTitle: the crystal structure of a putative pts iia(ptxa) from streptococcus2 mutans
Phyre2
| 8 |
|
PDB 2oqt chain D
Region: 1 - 144
Aligned: 138
Modelled: 142
Confidence: 100.0%
Identity: 20%
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein spy0176;
PDBTitle: structural genomics, the crystal structure of a putative2 pts iia domain from streptococcus pyogenes m1 gas
Phyre2
| 9 |
|
PDB 2a0j chain A
Region: 1 - 141
Aligned: 137
Modelled: 138
Confidence: 99.9%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:pts system, nitrogen regulatory iia protein;
PDBTitle: crystal structure of nitrogen regulatory protein iia-ntr from2 neisseria meningitidis
Phyre2
| 10 |
|
PDB 1a6j chain A
Region: 1 - 149
Aligned: 142
Modelled: 149
Confidence: 99.9%
Identity: 18%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 11 |
|
PDB 1hyn chain Q
Region: 63 - 148
Aligned: 84
Modelled: 86
Confidence: 96.9%
Identity: 11%
PDB header:membrane protein
Chain: Q: PDB Molecule:band 3 anion transport protein;
PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
Phyre2
| 12 |
|
PDB 1hyn chain P
Region: 63 - 148
Aligned: 84
Modelled: 86
Confidence: 96.9%
Identity: 11%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: Anion transport protein, cytoplasmic domain
Phyre2
| 13 |
|
PDB 2jya chain A
Region: 5 - 25
Aligned: 21
Modelled: 21
Confidence: 7.6%
Identity: 5%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1810;
PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
Phyre2
| 14 |
|
PDB 1sxj chain B domain 1
Region: 13 - 42
Aligned: 30
Modelled: 30
Confidence: 7.3%
Identity: 7%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: DNA polymerase III clamp loader subunits, C-terminal domain
Phyre2
| 15 |
|
PDB 1vlf chain M domain 2
Region: 14 - 28
Aligned: 15
Modelled: 15
Confidence: 5.7%
Identity: 13%
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3
Family: Formate dehydrogenase/DMSO reductase, domains 1-3
Phyre2
| 16 |
|
PDB 1g8k chain A domain 2
Region: 14 - 28
Aligned: 15
Modelled: 15
Confidence: 5.5%
Identity: 0%
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3
Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3
Family: Formate dehydrogenase/DMSO reductase, domains 1-3
Phyre2