| 1 |
|
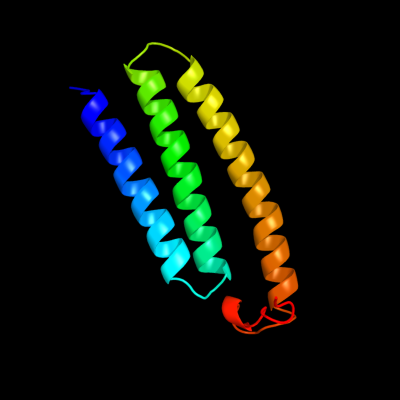
PDB 3rko chain K
Region: 1 - 100
Aligned: 100
Modelled: 100
Confidence: 100.0%
Identity: 100%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
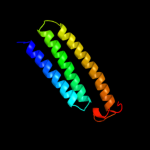
PDB 3rko chain L
Region: 5 - 98
Aligned: 91
Modelled: 94
Confidence: 79.7%
Identity: 22%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
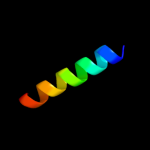
PDB 2jo1 chain A
Region: 8 - 26
Aligned: 19
Modelled: 19
Confidence: 54.0%
Identity: 47%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 4 |
|
PDB 2jp3 chain A
Region: 8 - 26
Aligned: 19
Modelled: 19
Confidence: 49.7%
Identity: 21%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 5 |
|
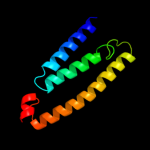
PDB 3rko chain M
Region: 1 - 91
Aligned: 91
Modelled: 91
Confidence: 41.2%
Identity: 13%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 2zxe chain G
Region: 8 - 26
Aligned: 19
Modelled: 19
Confidence: 40.6%
Identity: 42%
PDB header:hydrolase/transport protein
Chain: G: PDB Molecule:phospholemman-like protein;
PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
Phyre2
| 7 |
|
PDB 3rko chain N
Region: 7 - 91
Aligned: 82
Modelled: 85
Confidence: 25.2%
Identity: 22%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 8 |
|
PDB 1ifl chain A
Region: 49 - 89
Aligned: 41
Modelled: 41
Confidence: 24.8%
Identity: 10%
PDB header:virus
Chain: A: PDB Molecule:inovirus;
PDBTitle: molecular models and structural comparisons of native and2 mutant class i filamentous bacteriophages ff (fd, f1, m13),3 if1 and ike
Phyre2
| 9 |
|
PDB 3kdp chain H
Region: 8 - 26
Aligned: 19
Modelled: 19
Confidence: 22.5%
Identity: 47%
PDB header:hydrolase
Chain: H: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 10 |
|
PDB 3kdp chain G
Region: 8 - 26
Aligned: 19
Modelled: 19
Confidence: 22.5%
Identity: 47%
PDB header:hydrolase
Chain: G: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the sodium-potassium pump
Phyre2
| 11 |
|
PDB 1xme chain C domain 1
Region: 7 - 26
Aligned: 20
Modelled: 20
Confidence: 15.8%
Identity: 30%
Fold: Single transmembrane helix
Superfamily: Bacterial ba3 type cytochrome c oxidase subunit IIa
Family: Bacterial ba3 type cytochrome c oxidase subunit IIa
Phyre2
| 12 |
|
PDB 3n23 chain E
Region: 8 - 24
Aligned: 17
Modelled: 17
Confidence: 11.7%
Identity: 41%
PDB header:hydrolase
Chain: E: PDB Molecule:na+/k+ atpase gamma subunit transcript variant a;
PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
Phyre2
| 13 |
|
PDB 3mk7 chain F
Region: 62 - 94
Aligned: 33
Modelled: 33
Confidence: 10.4%
Identity: 9%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 14 |
|
PDB 3cto chain E
Region: 89 - 100
Aligned: 12
Modelled: 12
Confidence: 10.1%
Identity: 33%
PDB header:toxin inhibitor
Chain: E: PDB Molecule:uncharacterized protein rv3357/mt3465;
PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
Phyre2
| 15 |
|
PDB 1v54 chain M
Region: 68 - 84
Aligned: 17
Modelled: 17
Confidence: 8.5%
Identity: 24%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Family: Mitochondrial cytochrome c oxidase subunit VIIIb (aka IX)
Phyre2