| 1 |
|
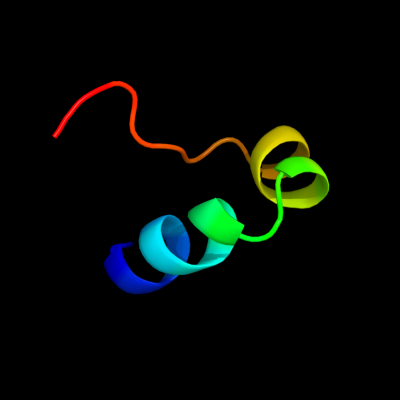
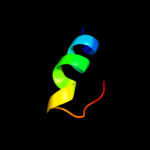
PDB 1pp7 chain U
Region: 108 - 132
Aligned: 25
Modelled: 25
Confidence: 54.5%
Identity: 24%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: 39 kda initiator binding protein, IBP39, N-terminal domain
Phyre2
| 2 |
|
PDB 2ga1 chain A domain 1
Region: 108 - 124
Aligned: 17
Modelled: 17
Confidence: 20.8%
Identity: 24%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Alr1493-like
Phyre2
| 3 |
|
PDB 3s81 chain A
Region: 86 - 121
Aligned: 36
Modelled: 36
Confidence: 12.8%
Identity: 8%
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
Phyre2
| 4 |
|
PDB 2i4c chain A
Region: 113 - 132
Aligned: 20
Modelled: 20
Confidence: 10.5%
Identity: 25%
PDB header:transport protein
Chain: A: PDB Molecule:bicarbonate transporter;
PDBTitle: crystal structure of bicarbonate transport protein cmpa from2 synechocystis sp. pcc 6803 in complex with bicarbonate and calcium
Phyre2
| 5 |
|
PDB 2fhw chain A
Region: 127 - 131
Aligned: 5
Modelled: 5
Confidence: 6.9%
Identity: 80%
PDB header:signaling protein
Chain: A: PDB Molecule:relaxin 3 (prorelaxin h3) (insulin-like peptide
PDBTitle: solution structure of human relaxin-3
Phyre2
| 6 |
|
PDB 2g29 chain A
Region: 113 - 132
Aligned: 20
Modelled: 20
Confidence: 6.7%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:nitrate transport protein nrta;
PDBTitle: crystal structure of the periplasmic nitrate-binding2 protein nrta from synechocystis pcc 6803
Phyre2
| 7 |
|
PDB 1sxj chain A domain 1
Region: 3 - 13
Aligned: 11
Modelled: 11
Confidence: 6.2%
Identity: 55%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: DNA polymerase III clamp loader subunits, C-terminal domain
Phyre2
| 8 |
|
PDB 2h7a chain A domain 1
Region: 20 - 34
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 20%
Fold: YcgL-like
Superfamily: YcgL-like
Family: YcgL-like
Phyre2
| 9 |
|
PDB 1xkp chain C domain 1
Region: 106 - 122
Aligned: 17
Modelled: 17
Confidence: 5.8%
Identity: 24%
Fold: Secretion chaperone-like
Superfamily: Type III secretory system chaperone-like
Family: Type III secretory system chaperone
Phyre2
| 10 |
|
PDB 1itw chain A
Region: 113 - 126
Aligned: 14
Modelled: 14
Confidence: 5.8%
Identity: 29%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Monomeric isocitrate dehydrogenase
Phyre2
| 11 |
|
PDB 2b0t chain A
Region: 113 - 126
Aligned: 14
Modelled: 14
Confidence: 5.3%
Identity: 21%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp isocitrate dehydrogenase;
PDBTitle: structure of monomeric nadp isocitrate dehydrogenase
Phyre2