| 1 |
|
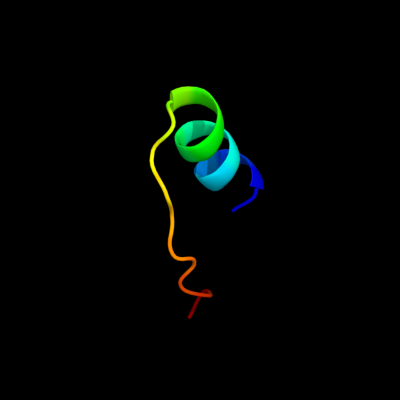
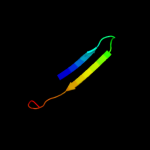
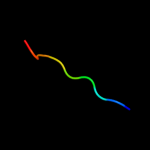
PDB 2jya chain A
Region: 67 - 87
Aligned: 21
Modelled: 21
Confidence: 25.8%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1810;
PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
Phyre2
| 2 |
|
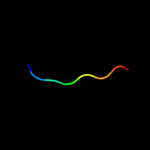
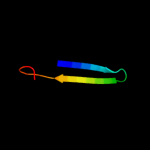
PDB 1dlj chain A domain 3
Region: 22 - 38
Aligned: 17
Modelled: 17
Confidence: 13.5%
Identity: 29%
Fold: Adenine nucleotide alpha hydrolase-like
Superfamily: UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
Family: UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
Phyre2
| 3 |
|
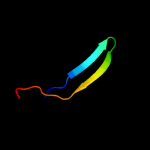
PDB 2h26 chain A domain 2
Region: 15 - 37
Aligned: 23
Modelled: 23
Confidence: 9.3%
Identity: 17%
Fold: MHC antigen-recognition domain
Superfamily: MHC antigen-recognition domain
Family: MHC antigen-recognition domain
Phyre2
| 4 |
|
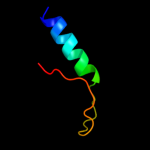
PDB 2po6 chain A domain 2
Region: 15 - 37
Aligned: 23
Modelled: 23
Confidence: 7.5%
Identity: 30%
Fold: MHC antigen-recognition domain
Superfamily: MHC antigen-recognition domain
Family: MHC antigen-recognition domain
Phyre2
| 5 |
|
PDB 2fik chain A domain 2
Region: 15 - 37
Aligned: 23
Modelled: 23
Confidence: 7.4%
Identity: 17%
Fold: MHC antigen-recognition domain
Superfamily: MHC antigen-recognition domain
Family: MHC antigen-recognition domain
Phyre2
| 6 |
|
PDB 1f32 chain A
Region: 17 - 28
Aligned: 12
Modelled: 12
Confidence: 7.1%
Identity: 33%
Fold: Pepsin inhibitor-3
Superfamily: Pepsin inhibitor-3
Family: Pepsin inhibitor-3
Phyre2
| 7 |
|
PDB 1x38 chain A domain 2
Region: 29 - 36
Aligned: 8
Modelled: 8
Confidence: 6.9%
Identity: 25%
Fold: Flavodoxin-like
Superfamily: Beta-D-glucan exohydrolase, C-terminal domain
Family: Beta-D-glucan exohydrolase, C-terminal domain
Phyre2
| 8 |
|
PDB 1k8i chain B
Region: 14 - 37
Aligned: 24
Modelled: 24
Confidence: 6.8%
Identity: 13%
PDB header:immune system
Chain: B: PDB Molecule:mhc class ii h2-m beta 2 chain;
PDBTitle: crystal structure of mouse h2-dm
Phyre2
| 9 |
|
PDB 2p24 chain A
Region: 17 - 37
Aligned: 21
Modelled: 21
Confidence: 6.7%
Identity: 19%
PDB header:immune system
Chain: A: PDB Molecule:h-2 class ii histocompatibility antigen, a-u alpha chain;
PDBTitle: i-au/mbp125-135
Phyre2
| 10 |
|
PDB 1vqo chain E domain 2
Region: 3 - 49
Aligned: 33
Modelled: 33
Confidence: 5.5%
Identity: 24%
Fold: Ribosomal protein L6
Superfamily: Ribosomal protein L6
Family: Ribosomal protein L6
Phyre2
| 11 |
|
PDB 1j0y chain D
Region: 97 - 105
Aligned: 9
Modelled: 9
Confidence: 5.3%
Identity: 44%
PDB header:hydrolase
Chain: D: PDB Molecule:beta-amylase;
PDBTitle: beta-amylase from bacillus cereus var. mycoides in complex2 with glucose
Phyre2
| 12 |
|
PDB 3dbx chain A
Region: 15 - 37
Aligned: 23
Modelled: 23
Confidence: 5.2%
Identity: 13%
PDB header:immune system
Chain: A: PDB Molecule:cd1-2 antigen;
PDBTitle: structure of chicken cd1-2 with bound fatty acid
Phyre2