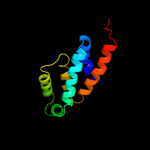
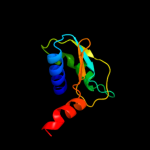
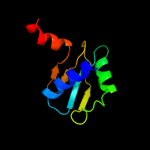
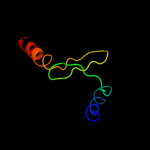
1 c2wssS_
100.0
26
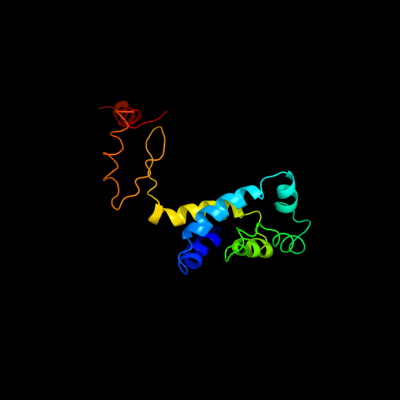
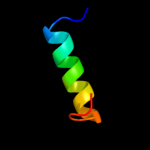
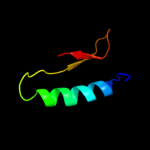
PDB header: hydrolaseChain: S: PDB Molecule: atp synthase subunit o, mitochondrial;PDBTitle: the structure of the membrane extrinsic region of bovine2 atp synthase
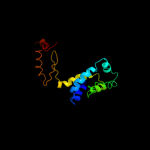
2 d1abva_
99.9
100
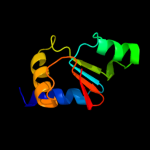
Fold: ATPD N-terminal domain-likeSuperfamily: N-terminal domain of the delta subunit of the F1F0-ATP synthaseFamily: N-terminal domain of the delta subunit of the F1F0-ATP synthase3 c2a7uB_
99.9
100
PDB header: hydrolaseChain: B: PDB Molecule: atp synthase delta chain;PDBTitle: nmr solution structure of the e.coli f-atpase delta subunit n-terminal2 domain in complex with alpha subunit n-terminal 22 residues
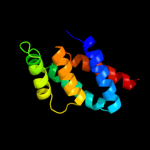
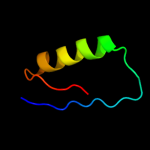
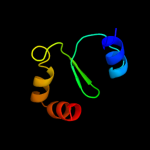
4 c2jmxA_
99.8
23
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase o subunit, mitochondrial;PDBTitle: oscp-nt (1-120) in complex with n-terminal (1-25) alpha2 subunit from f1-atpase
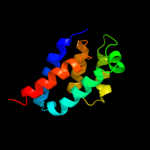
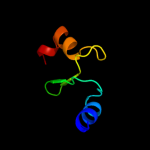
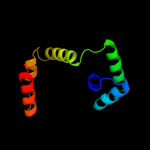
5 c2dm9B_
93.8
22
PDB header: hydrolaseChain: B: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of ph1978 from pyrococcus horikoshii ot3
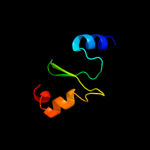
6 d2dm9a1
93.7
21
Fold: FwdE/GAPDH domain-likeSuperfamily: V-type ATPase subunit E-likeFamily: V-type ATPase subunit E7 c3lg8B_
85.1
22
PDB header: hydrolaseChain: B: PDB Molecule: a-type atp synthase subunit e;PDBTitle: crystal structure of the c-terminal part of subunit e (e101-206) from2 methanocaldococcus jannaschii of a1ao atp synthase
8 c3k5bE_
78.6
25
PDB header: hydrolaseChain: E: PDB Molecule: v-type atp synthase subunit e;PDBTitle: crystal structure of the peripheral stalk of thermus thermophilus h+-2 atpase/synthase
9 c3ipzA_
46.1
10
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-s14, chloroplastic;PDBTitle: crystal structure of arabidopsis monothiol glutaredoxin atgrxcp
10 c2ayaA_
29.6
15
PDB header: transferaseChain: A: PDB Molecule: dna polymerase iii subunit tau;PDBTitle: solution structure of the c-terminal 14 kda domain of the2 tau subunit from escherichia coli dna polymerase iii
11 c2jacA_
24.0
10
PDB header: electron transportChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: glutaredoxin grx1p c30s mutant from yeast
12 c3gx8A_
22.5
14
PDB header: electron transportChain: A: PDB Molecule: monothiol glutaredoxin-5, mitochondrial;PDBTitle: structural and biochemical characterization of yeast2 monothiol glutaredoxin grx5
13 d1sqwa2
20.9
14
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: Nip7p homolog, N-terminal domain14 c3c1sA_
19.9
10
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structure of grx1 in glutathionylated form
15 d2uubc1
19.5
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)16 c3d5jB_
15.8
20
PDB header: oxidoreductaseChain: B: PDB Molecule: glutaredoxin-2, mitochondrial;PDBTitle: structure of yeast grx2-c30s mutant with glutathionyl mixed2 disulfide
17 d2hc5a1
12.9
20
Fold: FlaG-likeSuperfamily: FlaG-likeFamily: FlaG-like18 c2klxA_
12.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: solution structure of glutaredoxin from bartonella henselae str.2 houston
19 d1okra_
10.5
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Penicillinase repressor20 c3h8qB_
9.9
20
PDB header: oxidoreductaseChain: B: PDB Molecule: thioredoxin reductase 3;PDBTitle: crystal structure of glutaredoxin domain of human thioredoxin2 reductase 3
21 d1ktea_
not modelled
9.5
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase22 c2rrnA_
not modelled
9.3
12
PDB header: protein transportChain: A: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: solution structure of secdf periplasmic domain p4
23 c2hzfA_
not modelled
9.0
14
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structures of a poxviral glutaredoxin in the oxidized and2 reduced states show redox-correlated structural changes
24 d1ts9a_
not modelled
8.8
14
Fold: Rof/RNase P subunit-likeSuperfamily: Rof/RNase P subunit-likeFamily: RNase P subunit p29-like25 d1w5fa2
not modelled
7.9
10
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain26 c1sqwA_
not modelled
7.7
14
PDB header: unknown functionChain: A: PDB Molecule: saccharomyces cerevisiae nip7p homolog;PDBTitle: crystal structure of kd93, a novel protein expressed in the2 human pro
27 c2q2gA_
not modelled
7.1
25
PDB header: chaperoneChain: A: PDB Molecule: heat shock 40 kda protein, putative (fragment);PDBTitle: crystal structure of dimerization domain of hsp40 from2 cryptosporidium parvum, cgd2_1800
28 c2ht9A_
not modelled
6.7
10
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin-2;PDBTitle: the structure of dimeric human glutaredoxin 2
29 c3ic4A_
not modelled
6.6
12
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin (grx-1);PDBTitle: the crystal structure of the glutaredoxin(grx-1) from archaeoglobus2 fulgidus
30 c2nn6E_
not modelled
6.5
20
PDB header: hydrolase/transferaseChain: E: PDB Molecule: exosome complex exonuclease rrp42;PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
31 c2kwaA_
not modelled
6.5
26
PDB header: transferase inhibitorChain: A: PDB Molecule: kinase a inhibitor;PDBTitle: 1h, 13c and 15n backbone and side chain resonance assignments of the2 n-terminal domain of the histidine kinase inhibitor kipi from3 bacillus subtilis
32 c2d4gA_
not modelled
6.3
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein bsu11850;PDBTitle: structure of yjcg protein, a putative 2'-5' rna ligase from2 bacillus subtilis
33 c3fzaA_
not modelled
6.1
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin;PDBTitle: crystal structure of poplar glutaredoxin s12 in complex with2 glutathione and beta-mercaptoethanol
34 c3qmxA_
not modelled
6.1
18
PDB header: electron transportChain: A: PDB Molecule: glutaredoxin a;PDBTitle: x-ray crystal structure of synechocystis sp. pcc 6803 glutaredoxin a
35 d1jhba_
not modelled
6.0
14
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase36 c2e7pC_
not modelled
5.7
17
PDB header: electron transportChain: C: PDB Molecule: glutaredoxin;PDBTitle: crystal structure of the holo form of glutaredoxin c1 from populus2 tremula x tremuloides
37 c2zdiA_
not modelled
5.7
13
PDB header: chaperoneChain: A: PDB Molecule: prefoldin subunit beta;PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
38 d1h72c1
not modelled
5.7
12
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain39 d1iqca2
not modelled
5.5
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase40 c3pp5A_
not modelled
5.3
31
PDB header: structural proteinChain: A: PDB Molecule: brk1;PDBTitle: high-resolution structure of the trimeric scar/wave complex precursor2 brk1
41 d1l8na1
not modelled
5.3
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: alpha-D-glucuronidase/Hyaluronidase catalytic domain42 d2ff4a1
not modelled
5.3
0
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: C-terminal effector domain of the bipartite response regulatorsFamily: PhoB-like43 c2ve7C_
not modelled
5.2
13
PDB header: cell cycleChain: C: PDB Molecule: kinetochore protein nuf2, kinetochore protein spc24;PDBTitle: crystal structure of a bonsai version of the human ndc802 complex
44 c2vhdB_
not modelled
5.1
13
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: crystal structure of the di-haem cytochrome c peroxidase2 from pseudomonas aeruginosa - mixed valence form