| 1 |
|
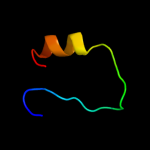
PDB 1sg5 chain A domain 1
Region: 32 - 42
Aligned: 11
Modelled: 11
Confidence: 24.5%
Identity: 18%
Fold: Rof/RNase P subunit-like
Superfamily: Rof/RNase P subunit-like
Family: Rof-like
Phyre2
| 2 |
|
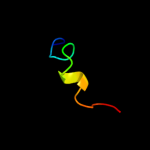
PDB 1h3g chain A domain 1
Region: 28 - 42
Aligned: 15
Modelled: 14
Confidence: 12.3%
Identity: 20%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: E-set domains of sugar-utilizing enzymes
Phyre2
| 3 |
|
PDB 3af5 chain A
Region: 53 - 79
Aligned: 27
Modelled: 27
Confidence: 9.1%
Identity: 37%
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein ph1404;
PDBTitle: the crystal structure of an archaeal cpsf subunit, ph1404 from2 pyrococcus horikoshii
Phyre2
| 4 |
|
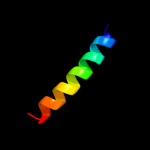
PDB 2a93 chain A
Region: 79 - 90
Aligned: 12
Modelled: 12
Confidence: 6.8%
Identity: 50%
PDB header:leucine zippers
Chain: A: PDB Molecule:c-myc-max heterodimeric leucine zipper;
PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, 40 structures
Phyre2
| 5 |
|
PDB 1a93 chain A
Region: 79 - 90
Aligned: 12
Modelled: 12
Confidence: 6.8%
Identity: 50%
PDB header:leucine zipper
Chain: A: PDB Molecule:myc proto-oncogene protein;
PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, nmr, minimized average structure
Phyre2
| 6 |
|
PDB 2h80 chain A domain 1
Region: 41 - 58
Aligned: 16
Modelled: 18
Confidence: 6.0%
Identity: 25%
Fold: SAM domain-like
Superfamily: SAM/Pointed domain
Family: Variant SAM domain
Phyre2
| 7 |
|
PDB 2oa5 chain A domain 1
Region: 70 - 92
Aligned: 23
Modelled: 23
Confidence: 5.9%
Identity: 22%
Fold: BLRF2-like
Superfamily: BLRF2-like
Family: BLRF2-like
Phyre2