| 1 |
|
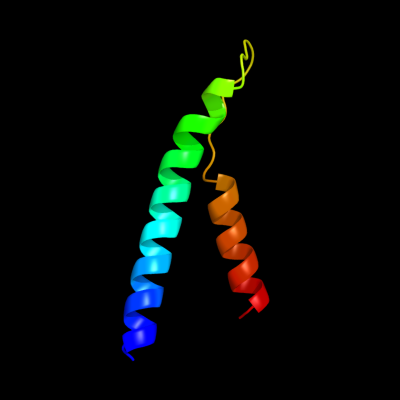
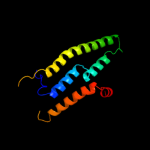
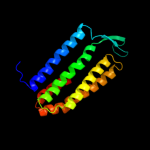
PDB 1kf6 chain C
Region: 127 - 180
Aligned: 54
Modelled: 54
Confidence: 22.2%
Identity: 19%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 2 |
|
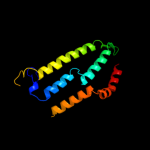
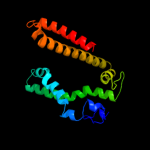
PDB 3ddl chain B
Region: 19 - 179
Aligned: 158
Modelled: 161
Confidence: 19.0%
Identity: 10%
PDB header:transport protein
Chain: B: PDB Molecule:xanthorhodopsin;
PDBTitle: crystallographic structure of xanthorhodopsin, a light-2 driven ion pump with dual chromophore
Phyre2
| 3 |
|
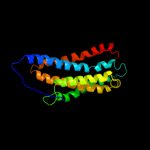
PDB 1h2s chain A
Region: 19 - 175
Aligned: 143
Modelled: 157
Confidence: 16.5%
Identity: 11%
Fold: Family A G protein-coupled receptor-like
Superfamily: Family A G protein-coupled receptor-like
Family: Bacteriorhodopsin-like
Phyre2
| 4 |
|
PDB 3a7k chain D
Region: 19 - 174
Aligned: 151
Modelled: 156
Confidence: 11.3%
Identity: 9%
PDB header:membrane protein
Chain: D: PDB Molecule:halorhodopsin;
PDBTitle: crystal structure of halorhodopsin from natronomonas2 pharaonis
Phyre2
| 5 |
|
PDB 3rko chain N
Region: 13 - 134
Aligned: 114
Modelled: 122
Confidence: 11.2%
Identity: 17%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 6 |
|
PDB 3rko chain M
Region: 9 - 146
Aligned: 126
Modelled: 138
Confidence: 10.7%
Identity: 12%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 7 |
|
PDB 2vay chain B
Region: 141 - 161
Aligned: 21
Modelled: 20
Confidence: 10.7%
Identity: 24%
PDB header:metal transport
Chain: B: PDB Molecule:voltage-dependent l-type calcium channel subunit
PDBTitle: calmodulin complexed with cav1.1 iq peptide
Phyre2
| 8 |
|
PDB 1pw4 chain A
Region: 9 - 193
Aligned: 181
Modelled: 185
Confidence: 10.4%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 9 |
|
PDB 1eys chain H domain 2
Region: 140 - 161
Aligned: 22
Modelled: 22
Confidence: 8.1%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 168 - 192
Aligned: 25
Modelled: 25
Confidence: 7.9%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 2kb1 chain A
Region: 54 - 106
Aligned: 49
Modelled: 53
Confidence: 7.5%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 12 |
|
PDB 2axt chain Z domain 1
Region: 135 - 186
Aligned: 52
Modelled: 52
Confidence: 6.5%
Identity: 21%
Fold: Transmembrane helix hairpin
Superfamily: PsbZ-like
Family: PsbZ-like
Phyre2
| 13 |
|
PDB 2jag chain A
Region: 19 - 174
Aligned: 151
Modelled: 156
Confidence: 5.9%
Identity: 6%
PDB header:membrane protein
Chain: A: PDB Molecule:halorhodopsin;
PDBTitle: l1-intermediate of halorhodopsin t203v
Phyre2
| 14 |
|
PDB 2k1h chain A
Region: 10 - 21
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 0%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ser13;
PDBTitle: solution nmr structure of ser13 from staphylococcus epidermidis.2 northeast structural genomics consortium target ser13
Phyre2
| 15 |
|
PDB 2xq2 chain A
Region: 10 - 182
Aligned: 173
Modelled: 173
Confidence: 5.5%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2