| 1 |
|
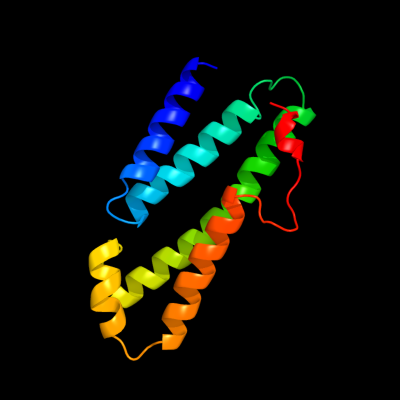
PDB 3rko chain L
Region: 95 - 220
Aligned: 126
Modelled: 126
Confidence: 90.1%
Identity: 17%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
PDB 2jpm chain A
Region: 51 - 81
Aligned: 31
Modelled: 31
Confidence: 28.8%
Identity: 23%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin lactococcin-g subunit beta;
PDBTitle: lactococcin g-b in tfe
Phyre2
| 3 |
|
PDB 1xn8 chain A
Region: 212 - 246
Aligned: 35
Modelled: 35
Confidence: 24.2%
Identity: 14%
Fold: Hypothetical protein YqbG
Superfamily: Hypothetical protein YqbG
Family: Hypothetical protein YqbG
Phyre2
| 4 |
|
PDB 3rko chain N
Region: 95 - 283
Aligned: 173
Modelled: 189
Confidence: 16.8%
Identity: 14%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 5 |
|
PDB 1q2i chain A
Region: 61 - 70
Aligned: 10
Modelled: 10
Confidence: 10.3%
Identity: 30%
PDB header:antitumor protein
Chain: A: PDB Molecule:pnc27;
PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
Phyre2
| 6 |
|
PDB 1jb0 chain L
Region: 140 - 246
Aligned: 85
Modelled: 87
Confidence: 6.7%
Identity: 20%
Fold: Photosystem I reaction center subunit XI, PsaL
Superfamily: Photosystem I reaction center subunit XI, PsaL
Family: Photosystem I reaction center subunit XI, PsaL
Phyre2
| 7 |
|
PDB 2dbh chain A
Region: 36 - 63
Aligned: 22
Modelled: 22
Confidence: 5.8%
Identity: 36%
PDB header:signaling protein
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily
PDBTitle: solution structure of the carboxyl-terminal card-like2 domain in human tnfr-related death receptor-6
Phyre2