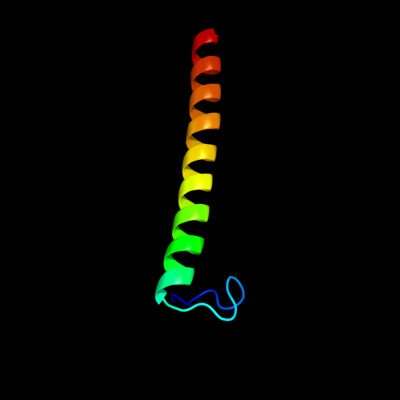
1 d1u7la_
37.4
17
Fold: Vacuolar ATP synthase subunit CSuperfamily: Vacuolar ATP synthase subunit CFamily: Vacuolar ATP synthase subunit C2 c2ahpB_
32.5
50
PDB header: de novo proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper, mutation of lys15 to epsilon-azido-lys
3 c2ahpA_
32.1
50
PDB header: de novo proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper, mutation of lys15 to epsilon-azido-lys
4 c3etoB_
27.3
24
PDB header: signaling proteinChain: B: PDB Molecule: neurogenic locus notch homolog protein 1;PDBTitle: 2 angstrom xray structure of the notch1 negative regulatory region2 (nrr)
5 c1ce0B_
26.3
35
PDB header: hiv-1 envelope proteinChain: B: PDB Molecule: protein (leucine zipper model h38-p1);PDBTitle: trimerization specificity in hiv-1 gp41: analysis with a2 gcn4 leucine zipper model
6 c1ij2C_
21.9
41
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
7 c1ij1B_
21.9
58
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvlt coiled-coil trimer with threonine at the d(12)2 position
8 c1ij1C_
21.9
58
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvlt coiled-coil trimer with threonine at the d(12)2 position
9 c1ij1A_
21.9
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvlt coiled-coil trimer with threonine at the d(12)2 position
10 c3k7zA_
21.8
41
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
11 c1rb1A_
21.8
41
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
12 c1rb6C_
21.8
41
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal form
13 c1swiA_
21.8
41
PDB header: leucine zipperChain: A: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
14 c1rb1B_
21.8
41
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
15 c3k7zB_
21.8
41
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
16 c1ij3C_
21.8
41
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
17 c1ij3B_
21.8
41
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
18 c2wpzA_
21.3
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
19 c1ij0B_
21.2
58
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: coiled coil trimer gcn4-pvls ser at buried d position
20 c1ij0A_
21.2
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: coiled coil trimer gcn4-pvls ser at buried d position
21 c1ij0C_
not modelled
21.2
58
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: coiled coil trimer gcn4-pvls ser at buried d position
22 c2wq1A_
not modelled
20.5
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating bromide
23 c2wq3A_
not modelled
20.5
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating chloride and nitrate
24 c2wpzB_
not modelled
20.1
58
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
25 c2wpzC_
not modelled
19.8
58
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with two vxxnxxx motifs2 coordinating chloride
26 c2o7hF_
not modelled
19.1
36
PDB header: transcriptionChain: F: PDB Molecule: general control protein gcn4;PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
27 c1ij2B_
not modelled
19.1
41
PDB header: transcriptionChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
28 c2dalA_
not modelled
19.0
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein kiaa0794;PDBTitle: solution structure of the novel identified uba-like domain2 in the n-terminal of human fas associated factor 1 protein
29 c1piqA_
not modelled
18.4
50
PDB header: dna binding proteinChain: A: PDB Molecule: protein (general control protein gcn4-piq);PDBTitle: crystal structure of gcn4-piq, a trimeric coiled coil with buried2 polar residues
30 c1zilA_
not modelled
17.7
45
PDB header: leucine zipperChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16gln in the dimeric2 state
31 c1zilB_
not modelled
17.7
45
PDB header: leucine zipperChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16gln in the dimeric2 state
32 c1rb4C_
not modelled
17.3
58
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal automatic solution
33 c1rb4A_
not modelled
16.6
58
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal automatic solution
34 c1swiB_
not modelled
16.6
58
PDB header: leucine zipperChain: B: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
35 c1ij2A_
not modelled
16.6
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvtl coiled-coil trimer with threonine at the a(16)2 position
36 c2ztaB_
not modelled
16.5
58
PDB header: leucine zipperChain: B: PDB Molecule: gcn4 leucine zipper;PDBTitle: x-ray structure of the gcn4 leucine zipper, a two-stranded,2 parallel coiled coil
37 c2ztaA_
not modelled
16.5
58
PDB header: leucine zipperChain: A: PDB Molecule: gcn4 leucine zipper;PDBTitle: x-ray structure of the gcn4 leucine zipper, a two-stranded,2 parallel coiled coil
38 c1gcmA_
not modelled
16.3
58
PDB header: transcription regulationChain: A: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
39 c3h25A_
not modelled
16.3
25
PDB header: replication/dnaChain: A: PDB Molecule: replication protein b;PDBTitle: crystal structure of the catalytic domain of primase repb' in complex2 with initiator dna
40 c1zimC_
not modelled
16.1
45
PDB header: leucine zipperChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16gln in the trimeric2 state
41 c1zimB_
not modelled
16.1
45
PDB header: leucine zipperChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16gln in the trimeric2 state
42 c2xzrA_
not modelled
15.9
37
PDB header: cell adhesionChain: A: PDB Molecule: immunoglobulin-binding protein eibd;PDBTitle: escherichia coli immunoglobulin-binding protein eibd 391-438 fused2 to gcn4 adaptors
43 d1xhmb1
not modelled
15.8
33
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Transducin (heterotrimeric G protein), gamma chainFamily: Transducin (heterotrimeric G protein), gamma chain44 c1zikA_
not modelled
15.8
58
PDB header: leucine zipperChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16lys in the dimeric2 state
45 c1zikB_
not modelled
15.8
58
PDB header: leucine zipperChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16lys in the dimeric2 state
46 c1ce9C_
not modelled
15.8
64
PDB header: helix cappingChain: C: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
47 c1ce9D_
not modelled
15.8
64
PDB header: helix cappingChain: D: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
48 c1ce9A_
not modelled
15.8
64
PDB header: helix cappingChain: A: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
49 c1ce9B_
not modelled
15.8
64
PDB header: helix cappingChain: B: PDB Molecule: protein (gcn4-pmse);PDBTitle: helix capping in the gcn4 leucine zipper
50 c1xhmB_
not modelled
15.5
33
PDB header: signaling proteinChain: B: PDB Molecule: guanine nucleotide-binding protein g(i)/g(s)PDBTitle: the crystal structure of a biologically active peptide2 (sigk) bound to a g protein beta:gamma heterodimer
51 c1ld4E_
not modelled
15.5
64
PDB header: virusChain: E: PDB Molecule: general control protein gcn4;PDBTitle: placement of the structural proteins in sindbis virus
52 c2wpyA_
not modelled
15.4
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with one vxxnxxx motif2 coordinating chloride
53 c1ij3A_
not modelled
15.2
58
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-pvsl coiled-coil trimer with serine at the a(16)2 position
54 c1rb4B_
not modelled
15.2
58
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal automatic solution
55 c1rb6B_
not modelled
15.2
58
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal form
56 c1rb5C_
not modelled
15.2
58
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a trigonal form
57 c1rb5B_
not modelled
14.8
58
PDB header: dna binding proteinChain: B: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a trigonal form
58 c1rb6A_
not modelled
14.8
58
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a tetragonal form
59 c1rb5A_
not modelled
14.8
58
PDB header: dna binding proteinChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel trimer of gcn4-leucine zipper core mutant as2 n16a trigonal form
60 c1gcmB_
not modelled
14.5
58
PDB header: transcription regulationChain: B: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
61 c1gcmC_
not modelled
14.1
58
PDB header: transcription regulationChain: C: PDB Molecule: gcn4p-ii;PDBTitle: gcn4 leucine zipper core mutant p-li
62 c1ziiA_
not modelled
14.1
44
PDB header: leucine zipperChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16aba in the dimeric2 state
63 c1ziiB_
not modelled
14.1
44
PDB header: leucine zipperChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16aba in the dimeric2 state
64 d1pzra_
not modelled
14.0
45
Fold: HLH-likeSuperfamily: Docking domain B of the erythromycin polyketide synthase (DEBS)Family: Docking domain B of the erythromycin polyketide synthase (DEBS)65 c1zimA_
not modelled
13.9
48
PDB header: leucine zipperChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16gln in the trimeric2 state
66 c3p8mD_
not modelled
13.8
58
PDB header: protein bindingChain: D: PDB Molecule: general control protein gcn4;PDBTitle: human dynein light chain (dynll2) in complex with an in vitro evolved2 peptide dimerized by leucine zipper
67 c3k7zC_
not modelled
13.6
64
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
68 c1rb1C_
not modelled
13.6
64
PDB header: dna binding proteinChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant as n16a trigonal automatic2 solution
69 d1qm4a2
not modelled
13.6
31
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase70 d1mxaa2
not modelled
13.1
33
Fold: S-adenosylmethionine synthetaseSuperfamily: S-adenosylmethionine synthetaseFamily: S-adenosylmethionine synthetase71 c1ztaA_
not modelled
12.7
42
PDB header: dna-binding motifChain: A: PDB Molecule: leucine zipper monomer;PDBTitle: the solution structure of a leucine-zipper motif peptide
72 c2r9iA_
not modelled
12.6
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative phage capsid protein;PDBTitle: crystal structure of putative phage capsid protein domain from2 corynebacterium diphtheriae
73 c1zijB_
not modelled
12.4
44
PDB header: leucine zipperChain: B: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16aba in the trimeric2 state
74 c1zijA_
not modelled
12.4
44
PDB header: leucine zipperChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16aba in the trimeric2 state
75 c1zijC_
not modelled
12.4
44
PDB header: leucine zipperChain: C: PDB Molecule: general control protein gcn4;PDBTitle: gcn4-leucine zipper core mutant asn16aba in the trimeric2 state
76 c1swiC_
not modelled
12.0
64
PDB header: leucine zipperChain: C: PDB Molecule: gcn4p1;PDBTitle: gcn4-leucine zipper core mutant as n16a complexed with2 benzene
77 c1qeyA_
not modelled
12.0
50
PDB header: gene regulationChain: A: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
78 c1qeyB_
not modelled
12.0
50
PDB header: gene regulationChain: B: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
79 c1qeyC_
not modelled
12.0
50
PDB header: gene regulationChain: C: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
80 c1qeyD_
not modelled
12.0
50
PDB header: gene regulationChain: D: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
81 c3e9eB_
not modelled
11.7
18
PDB header: hydrolaseChain: B: PDB Molecule: zgc:56074;PDBTitle: structure of full-length h11a mutant form of tigar from danio rerio
82 c2wq2A_
not modelled
11.4
78
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating iodide
83 c2ccfA_
not modelled
10.5
50
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: antiparallel configuration of pli e20s
84 c2wq0A_
not modelled
10.3
78
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: gcn4 leucine zipper mutant with three ixxntxx motifs2 coordinating chloride
85 c1u9fA_
not modelled
10.2
50
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)
86 c1unwB_
not modelled
9.8
50
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
87 c1unxA_
not modelled
9.7
50
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
88 c1unyB_
not modelled
9.7
50
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
89 c2cceA_
not modelled
9.7
50
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: parallel configuration of pli e20s
90 c2cceB_
not modelled
9.7
50
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: parallel configuration of pli e20s
91 c1uo0A_
not modelled
9.6
50
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
92 c1uo0B_
not modelled
9.6
50
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
93 c1uo1B_
not modelled
9.6
50
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
94 c1u9hA_
not modelled
9.6
50
PDB header: transcriptionChain: A: PDB Molecule: general control protein gcn4;PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of e(22)l(23)
95 c1gclA_
not modelled
9.5
50
PDB header: leucine zipperChain: A: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
96 c1gclC_
not modelled
9.5
50
PDB header: leucine zipperChain: C: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
97 c3f1bA_
not modelled
9.3
10
PDB header: transcription regulatorChain: A: PDB Molecule: tetr-like transcriptional regulator;PDBTitle: the crystal structure of a tetr-like transcriptional regulator from2 rhodococcus sp. rha1.
98 c1gclB_
not modelled
9.2
50
PDB header: leucine zipperChain: B: PDB Molecule: gcn4;PDBTitle: gcn4 leucine zipper core mutant p-li
99 c1u9fC_
not modelled
9.1
50
PDB header: transcriptionChain: C: PDB Molecule: general control protein gcn4;PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(15)l(16)