| 1 |
|
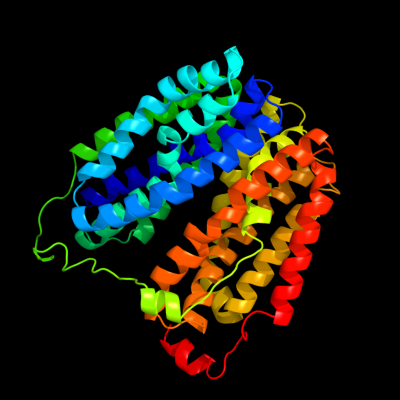
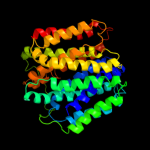
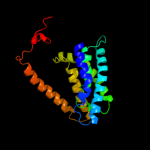
PDB 1pv7 chain A
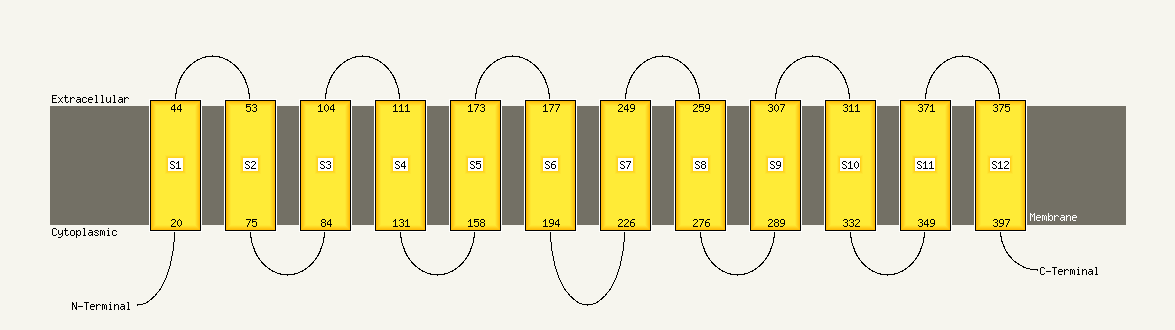
Region: 15 - 413
Aligned: 399
Modelled: 399
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 2 |
|
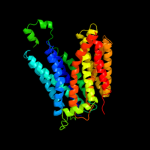
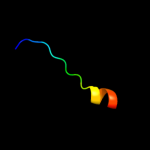
PDB 1pw4 chain A
Region: 3 - 410
Aligned: 406
Modelled: 408
Confidence: 100.0%
Identity: 12%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
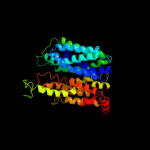
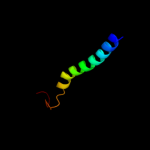
PDB 2gfp chain A
Region: 21 - 390
Aligned: 364
Modelled: 370
Confidence: 100.0%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
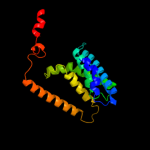
PDB 3o7p chain A
Region: 17 - 401
Aligned: 381
Modelled: 385
Confidence: 100.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 22 - 397
Aligned: 372
Modelled: 376
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 2jo1 chain A
Region: 371 - 415
Aligned: 45
Modelled: 45
Confidence: 16.0%
Identity: 16%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 7 |
|
PDB 2g9p chain A
Region: 71 - 84
Aligned: 14
Modelled: 14
Confidence: 13.7%
Identity: 29%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 8 |
|
PDB 3hd6 chain A
Region: 25 - 219
Aligned: 195
Modelled: 195
Confidence: 8.0%
Identity: 10%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 9 |
|
PDB 3hur chain A
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 7.2%
Identity: 13%
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from oenococcus oeni
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 376 - 416
Aligned: 41
Modelled: 41
Confidence: 6.7%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 3b9y chain A
Region: 34 - 223
Aligned: 190
Modelled: 190
Confidence: 6.5%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 12 |
|
PDB 2jp3 chain A
Region: 371 - 415
Aligned: 45
Modelled: 45
Confidence: 6.4%
Identity: 18%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 13 |
|
PDB 1rh1 chain A domain 2
Region: 9 - 81
Aligned: 69
Modelled: 73
Confidence: 6.2%
Identity: 17%
Fold: Toxins' membrane translocation domains
Superfamily: Colicin
Family: Colicin
Phyre2
| 14 |
|
PDB 1xl6 chain B
Region: 1 - 102
Aligned: 102
Modelled: 102
Confidence: 5.4%
Identity: 8%
PDB header:metal transport
Chain: B: PDB Molecule:inward rectifier potassium channel;
PDBTitle: intermediate gating structure 2 of the inwardly rectifying k+ channel2 kirbac3.1
Phyre2