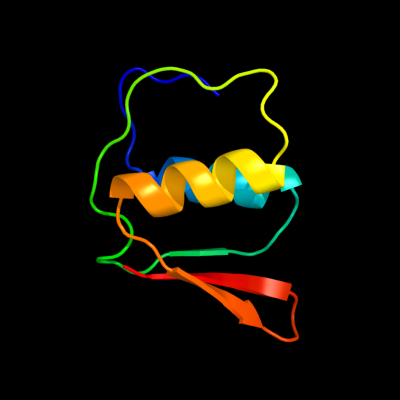
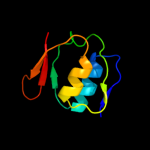
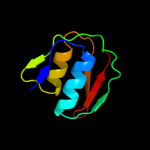
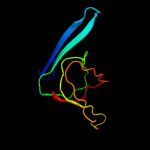
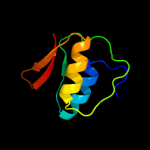
1 c2y9kG_
98.2
14
PDB header: protein transportChain: G: PDB Molecule: protein invg;PDBTitle: three-dimensional model of salmonella's needle complex at2 subnanometer resolution
2 c3gr5A_
97.9
9
PDB header: membrane proteinChain: A: PDB Molecule: escc;PDBTitle: periplasmic domain of the outer membrane secretin escc from2 enteropathogenic e.coli (epec)
3 c2y3mA_
96.2
14
PDB header: transport proteinChain: A: PDB Molecule: protein transport protein hofq;PDBTitle: structure of the extra-membranous domain of the secretin2 hofq from actinobacillus actinomycetemcomitans
4 c2a02A_
96.1
14
PDB header: membrane protein, metal transportChain: A: PDB Molecule: ferric-pseudobactin 358 receptor;PDBTitle: solution nmr structure of the periplasmic signaling domain2 of the outer membrane iron transporter pupa from3 pseudomonas putida.
5 c3ezjA_
96.1
10
PDB header: protein transportChain: A: PDB Molecule: general secretion pathway protein gspd;PDBTitle: crystal structure of the n-terminal domain of the secretin gspd from2 etec determined with the assistance of a nanobody
6 c3hugJ_
95.7
12
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
7 c3ossD_
95.4
17
PDB header: protein transportChain: D: PDB Molecule: type 2 secretion system, secretin gspd;PDBTitle: the crystal structure of enterotoxigenic escherichia coli gspc-gspd2 complex from the type ii secretion system
8 c2d1uA_
94.9
10
PDB header: metal transportChain: A: PDB Molecule: iron(iii) dicitrate transport protein feca;PDBTitle: solution strcuture of the periplasmic signaling domain of2 feca from escherichia coli
9 c1zzvA_
94.8
10
PDB header: membrane protein, metal transportChain: A: PDB Molecule: iron(iii) dicitrate transport protein feca;PDBTitle: solution nmr structure of the periplasmic signaling domain2 of the outer membrane iron transporter feca from3 escherichia coli.
10 c3eo6B_
86.1
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function (duf1255);PDBTitle: crystal structure of protein of unknown function (duf1255)2 (afe_2634) from acidithiobacillus ferrooxidans ncib8455 at3 0.97 a resolution
11 c3cddD_
85.2
14
PDB header: structural proteinChain: D: PDB Molecule: prophage muso2, 43 kda tail protein;PDBTitle: crystal structure of prophage muso2, 43 kda tail protein from2 shewanella oneidensis
12 c3hqxA_
85.1
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0345 protein aciad0356;PDBTitle: crystal structure of protein of unknown function (duf1255,pf06865)2 from acinetobacter sp. adp1
13 d3cdda2
79.1
15
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like14 c2z2sD_
78.7
8
PDB header: transcriptionChain: D: PDB Molecule: anti-sigma factor chrr, transcriptional activator chrr;PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
15 d1uika1
74.2
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein16 d1uija1
71.0
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein17 c2eaaB_
65.8
11
PDB header: plant proteinChain: B: PDB Molecule: 7s globulin-3;PDBTitle: crystal structure of adzuki bean 7s globulin-3
18 c3s7eB_
64.5
12
PDB header: allergenChain: B: PDB Molecule: allergen ara h 1, clone p41b;PDBTitle: crystal structure of ara h 1
19 d2bnma2
62.8
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1459-like20 c2pfwB_
61.2
7
PDB header: unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel domain protein;PDBTitle: crystal structure of a rmlc-like cupin (sfri_3105) from shewanella2 frigidimarina ncimb 400 at 1.90 a resolution
21 c2cauA_
not modelled
56.9
10
PDB header: plant proteinChain: A: PDB Molecule: protein (canavalin);PDBTitle: canavalin from jack bean
22 d1yhfa1
not modelled
56.1
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1287-like23 d1dgwa_
not modelled
56.0
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein24 c2iahA_
not modelled
54.7
14
PDB header: membrane proteinChain: A: PDB Molecule: ferripyoverdine receptor;PDBTitle: crystal structure of the ferripyoverdine receptor of the outer2 membrane of pseudomonas aeruginosa bound to ferripyoverdine.
25 d1fxza2
not modelled
51.7
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein26 c2bnoA_
not modelled
50.5
10
PDB header: oxidoreductaseChain: A: PDB Molecule: epoxidase;PDBTitle: the structure of hydroxypropylphosphonic acid epoxidase2 from s. wedmorenis.
27 c1uijA_
not modelled
46.9
8
PDB header: sugar binding proteinChain: A: PDB Molecule: beta subunit of beta conglycinin;PDBTitle: crystal structure of soybean beta-conglycinin beta2 homotrimer (i122m/k124w)
28 d2oyza1
not modelled
45.1
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: VPA0057-like29 d1tkea1
not modelled
45.1
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain30 c2vz1A_
not modelled
42.2
10
PDB header: oxidoreductaseChain: A: PDB Molecule: galactose oxidase;PDBTitle: premat-galactose oxidase
31 c3es4B_
not modelled
40.3
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf861 with a rmlc-like cupin fold;PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
32 c1wwtA_
not modelled
38.4
15
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
33 d1dw0a_
not modelled
38.0
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c34 d1yfua1
not modelled
36.7
8
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like35 d1y9qa2
not modelled
36.3
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Probable transcriptional regulator VC1968, C-terminal domain36 d2j73a1
not modelled
36.2
11
Fold: Prealbumin-likeSuperfamily: Starch-binding domain-likeFamily: PUD-like37 c2ozjB_
not modelled
34.8
12
PDB header: unknown functionChain: B: PDB Molecule: cupin 2, conserved barrel;PDBTitle: crystal structure of a cupin superfamily protein (dsy2733) from2 desulfitobacterium hafniense dcb-2 at 1.60 a resolution
38 c3myxA_
not modelled
32.8
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pspto_0244;PDBTitle: crystal structure of a pspto_0244 (protein with unknown function which2 belongs to pfam duf861 family) from pseudomonas syringae pv. tomato3 str. dc3000 at 1.30 a resolution
39 c3kscD_
not modelled
30.7
13
PDB header: plant proteinChain: D: PDB Molecule: lega class;PDBTitle: crystal structure of pea prolegumin, an 11s seed globulin2 from pisum sativum l.
40 d1j58a_
not modelled
30.4
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein41 d2phla1
not modelled
30.3
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein42 d1j1la_
not modelled
30.2
15
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Pirin-like43 c3d37A_
not modelled
30.0
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tail protein, 43 kda;PDBTitle: the crystal structure of the tail protein from neisseria meningitidis2 mc58
44 c3bcwB_
not modelled
29.6
9
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf861 family protein with a rmlc-like cupin2 fold (bb1179) from bordetella bronchiseptica rb50 at 1.60 a3 resolution
45 d1gu2a_
not modelled
28.7
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c46 d1v8qa_
not modelled
26.2
20
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: Ribosomal L27 protein47 d1o5ua_
not modelled
25.9
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111248 d1od5a2
not modelled
25.8
7
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein49 c1wruA_
not modelled
25.6
13
PDB header: structural proteinChain: A: PDB Molecule: 43 kda tail protein;PDBTitle: structure of central hub elucidated by x-ray analysis of gene product2 44; baseplate component of bacteriophage mu
50 d2pyta1
not modelled
25.2
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like51 d1wxma1
not modelled
25.1
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ras-binding domain, RBD52 c3o5cA_
not modelled
22.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: cytochrome c peroxidase bccp of shewanella oneidensis
53 d1nyra2
not modelled
22.6
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain54 d1wrua2
not modelled
21.8
13
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like55 c1fxzC_
not modelled
21.7
10
PDB header: plant proteinChain: C: PDB Molecule: glycinin g1;PDBTitle: crystal structure of soybean proglycinin a1ab1b homotrimer
56 c3kglB_
not modelled
21.7
15
PDB header: plant proteinChain: B: PDB Molecule: cruciferin;PDBTitle: crystal structure of procruciferin, 11s globulin from2 brassica napus
57 d1od5a1
not modelled
21.6
14
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein58 c2oziA_
not modelled
21.3
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rpa4178;PDBTitle: structural genomics, the crystal structure of a putative2 protein rpa4178 from rhodopseudomonas palustris cga009
59 c2gu9B_
not modelled
21.3
6
PDB header: immune systemChain: B: PDB Molecule: tetracenomycin polyketide synthesis protein;PDBTitle: crystal structure of xc5357 from xanthomonas campestris: a2 putative tetracenomycin polyketide synthesis protein3 adopting a novel cupin subfamily structure
60 d1vj2a_
not modelled
21.2
12
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1459-like61 c3ehkC_
not modelled
21.2
8
PDB header: plant proteinChain: C: PDB Molecule: prunin;PDBTitle: crystal structure of pru du amandin, an allergenic protein2 from prunus dulcis
62 d2oa4a1
not modelled
20.5
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: SPO1678-like63 c3hq7A_
not modelled
19.8
10
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: ccpa from g. sulfurreducens, g94k/k97q/r100i variant
64 c2c1uB_
not modelled
19.7
15
PDB header: oxidoreductaseChain: B: PDB Molecule: di-haem cytochrome c peroxidase;PDBTitle: crystal structure of the di-haem cytochrome c peroxidase2 from paracoccus pantotrophus - oxidised form
65 c3qacA_
not modelled
19.7
10
PDB header: plant proteinChain: A: PDB Molecule: 11s globulin seed storage protein;PDBTitle: structure of amaranth 11s proglobulin seed storage protein from2 amaranthus hypochondriacus l.
66 c3c3vA_
not modelled
19.5
10
PDB header: allergenChain: A: PDB Molecule: arachin arah3 isoform;PDBTitle: crystal structure of peanut major allergen ara h 3
67 c3rnsA_
not modelled
19.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cupin 2 conserved barrel domain protein;PDBTitle: cupin 2 conserved barrel domain protein from leptotrichia buccalis
68 d2f4pa1
not modelled
18.8
11
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: TM1287-like69 c2e9qA_
not modelled
18.7
8
PDB header: plant proteinChain: A: PDB Molecule: 11s globulin subunit beta;PDBTitle: recombinant pro-11s globulin of pumpkin
70 c2vecA_
not modelled
18.7
10
PDB header: cytosolic proteinChain: A: PDB Molecule: pirin-like protein yhak;PDBTitle: the crystal structure of the protein yhak from escherichia2 coli
71 c2d5fB_
not modelled
18.6
14
PDB header: plant proteinChain: B: PDB Molecule: glycinin a3b4 subunit;PDBTitle: crystal structure of recombinant soybean proglycinin a3b4 subunit, its2 comparison with mature glycinin a3b4 subunit, responsible for hexamer3 assembly
72 d1eb7a2
not modelled
18.0
13
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase73 c3pdkB_
not modelled
17.9
20
PDB header: isomeraseChain: B: PDB Molecule: phosphoglucosamine mutase;PDBTitle: crystal structure of phosphoglucosamine mutase from b. anthracis
74 d1mpga2
not modelled
16.9
10
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: DNA repair glycosylase, N-terminal domain75 c2i45C_
not modelled
16.4
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein nmb1881 from neisseria meningitidis
76 d2p5zx2
not modelled
16.3
7
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like77 c1nmlA_
not modelled
15.9
8
PDB header: oxidoreductaseChain: A: PDB Molecule: di-haem cytochrome c peroxidase;PDBTitle: di-haemic cytochrome c peroxidase from pseudomonas nautica 617, form2 in (ph 4.0)
78 d1fxza1
not modelled
15.9
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein79 c2jrtA_
not modelled
15.4
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr solution structure of the protein coded by gene2 rhos4_12090 of rhodobacter sphaeroides. northeast3 structural genomics target rhr5
80 c1y9qA_
not modelled
15.1
17
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, hth_3 family;PDBTitle: crystal structure of hth_3 family transcriptional regulator2 from vibrio cholerae
81 d1dlja3
not modelled
15.1
9
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: UDP-glucose/GDP-mannose dehydrogenase C-terminal domainFamily: UDP-glucose/GDP-mannose dehydrogenase C-terminal domain82 c3fjsC_
not modelled
14.9
16
PDB header: biosynthetic proteinChain: C: PDB Molecule: uncharacterized protein with rmlc-like cupin fold;PDBTitle: crystal structure of a putative biosynthetic protein with rmlc-like2 cupin fold (reut_b4087) from ralstonia eutropha jmp134 at 1.90 a3 resolution
83 d1nmla2
not modelled
14.9
10
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase84 c3ht2A_
not modelled
14.8
11
PDB header: lyaseChain: A: PDB Molecule: remf protein;PDBTitle: zink containing polyketide cyclase remf from streptomyces2 resistomycificus
85 c3fin0_
not modelled
14.3
20
PDB header: ribosomeChain: 0: PDB Molecule: 50s ribosomal protein l27;PDBTitle: t. thermophilus 70s ribosome in complex with mrna, trnas2 and ef-tu.gdp.kirromycin ternary complex, fitted to a 6.43 a cryo-em map. this file contains the 50s subunit.
86 c3i4oA_
not modelled
13.9
9
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-1;PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis
87 d1iqca2
not modelled
13.5
10
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase88 c1cauB_
not modelled
13.3
8
PDB header: seed storage proteinChain: B: PDB Molecule: canavalin;PDBTitle: determination of three crystal structures of canavalin by molecular2 replacement
89 d1pmia_
not modelled
13.3
6
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase90 c2qjvB_
not modelled
13.2
16
PDB header: isomeraseChain: B: PDB Molecule: uncharacterized iolb-like protein;PDBTitle: crystal structure of an iolb-like protein (stm4420) from salmonella2 typhimurium lt2 at 1.90 a resolution
91 d2bsya1
not modelled
12.8
8
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like92 d2zjrt1
not modelled
12.7
18
Fold: Barrel-sandwich hybridSuperfamily: Ribosomal L27 protein-likeFamily: Ribosomal L27 protein93 c1or7C_
not modelled
12.3
14
PDB header: transcriptionChain: C: PDB Molecule: sigma-e factor negative regulatory protein;PDBTitle: crystal structure of escherichia coli sigmae with the cytoplasmic2 domain of its anti-sigma rsea
94 d1or7c_
not modelled
12.3
14
Fold: N-terminal, cytoplasmic domain of anti-sigmaE factor RseASuperfamily: N-terminal, cytoplasmic domain of anti-sigmaE factor RseAFamily: N-terminal, cytoplasmic domain of anti-sigmaE factor RseA95 c2opkC_
not modelled
12.2
17
PDB header: isomeraseChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a putative mannose-6-phosphate isomerase2 (reut_a1446) from ralstonia eutropha jmp134 at 2.10 a resolution
96 c2vhdB_
not modelled
11.9
13
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c551 peroxidase;PDBTitle: crystal structure of the di-haem cytochrome c peroxidase2 from pseudomonas aeruginosa - mixed valence form
97 d1ekxa1
not modelled
11.9
14
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase98 d1ml4a1
not modelled
11.1
19
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase99 d1otha1
not modelled
10.9
19
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase