| 1 |
|
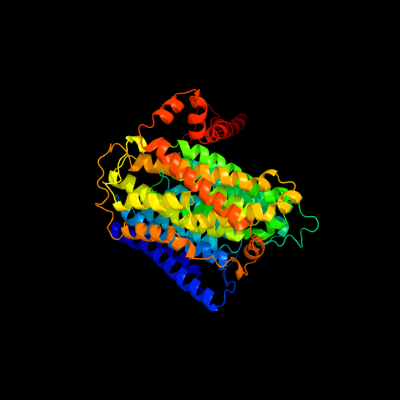
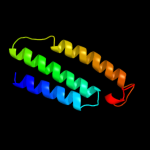
PDB 3rko chain L
Region: 4 - 592
Aligned: 546
Modelled: 575
Confidence: 100.0%
Identity: 21%
PDB header:oxidoreductase
Chain: L: PDB Molecule:nadh-quinone oxidoreductase subunit l;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
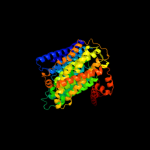
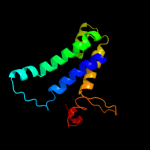
PDB 3rko chain M
Region: 6 - 489
Aligned: 461
Modelled: 484
Confidence: 100.0%
Identity: 21%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 3 |
|
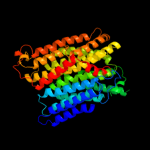
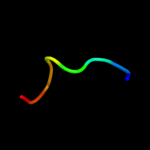
PDB 3rko chain N
Region: 1 - 488
Aligned: 456
Modelled: 487
Confidence: 100.0%
Identity: 18%
PDB header:oxidoreductase
Chain: N: PDB Molecule:nadh-quinone oxidoreductase subunit n;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 4 |
|
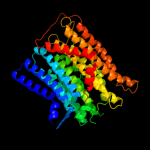
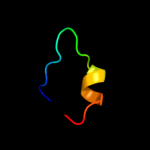
PDB 3rko chain K
Region: 107 - 186
Aligned: 80
Modelled: 80
Confidence: 33.2%
Identity: 16%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 5 |
|
PDB 2l3i chain A
Region: 278 - 289
Aligned: 12
Modelled: 12
Confidence: 24.2%
Identity: 42%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:aoxki4a, antimicrobial peptide in spider venom;
PDBTitle: oxki4a, spider derived antimicrobial peptide
Phyre2
| 6 |
|
PDB 1s1q chain A
Region: 212 - 227
Aligned: 16
Modelled: 16
Confidence: 24.0%
Identity: 19%
Fold: UBC-like
Superfamily: UBC-like
Family: UEV domain
Phyre2
| 7 |
|
PDB 1a6q chain A domain 1
Region: 367 - 400
Aligned: 34
Modelled: 34
Confidence: 19.3%
Identity: 12%
Fold: Another 3-helical bundle
Superfamily: Protein serine/threonine phosphatase 2C, C-terminal domain
Family: Protein serine/threonine phosphatase 2C, C-terminal domain
Phyre2
| 8 |
|
PDB 1o8b chain B domain 1
Region: 281 - 306
Aligned: 23
Modelled: 26
Confidence: 14.4%
Identity: 17%
Fold: NagB/RpiA/CoA transferase-like
Superfamily: NagB/RpiA/CoA transferase-like
Family: D-ribose-5-phosphate isomerase (RpiA), catalytic domain
Phyre2
| 9 |
|
PDB 3na2 chain C
Region: 217 - 228
Aligned: 12
Modelled: 12
Confidence: 10.5%
Identity: 25%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function from mine drainage2 metagenome leptospirillum rubarum
Phyre2
| 10 |
|
PDB 1j4n chain A
Region: 329 - 453
Aligned: 106
Modelled: 125
Confidence: 9.3%
Identity: 16%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 11 |
|
PDB 3d37 chain A domain 2
Region: 216 - 223
Aligned: 8
Modelled: 8
Confidence: 8.3%
Identity: 38%
Fold: Phage tail proteins
Superfamily: Phage tail proteins
Family: Baseplate protein-like
Phyre2
| 12 |
|
PDB 2yrk chain A domain 1
Region: 537 - 555
Aligned: 19
Modelled: 19
Confidence: 6.9%
Identity: 26%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: HkH motif-containing C2H2 finger
Phyre2
| 13 |
|
PDB 2c1h chain A domain 1
Region: 536 - 559
Aligned: 24
Modelled: 24
Confidence: 6.6%
Identity: 21%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
Phyre2
| 14 |
|
PDB 1l6s chain A
Region: 536 - 559
Aligned: 24
Modelled: 24
Confidence: 6.3%
Identity: 25%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
Phyre2
| 15 |
|
PDB 1gzg chain A
Region: 536 - 559
Aligned: 24
Modelled: 24
Confidence: 6.2%
Identity: 25%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
Phyre2
| 16 |
|
PDB 2q09 chain A domain 1
Region: 535 - 547
Aligned: 11
Modelled: 13
Confidence: 5.7%
Identity: 36%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: Imidazolonepropionase-like
Phyre2
| 17 |
|
PDB 2z84 chain A
Region: 535 - 539
Aligned: 5
Modelled: 5
Confidence: 5.4%
Identity: 80%
PDB header:hydrolase
Chain: A: PDB Molecule:ufm1-specific protease 1;
PDBTitle: insights from crystal and solution structures of mouse ufsp1
Phyre2
| 18 |
|
PDB 2imr chain A domain 1
Region: 530 - 542
Aligned: 13
Modelled: 13
Confidence: 5.4%
Identity: 38%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: DR0824-like
Phyre2