| 1 |
|
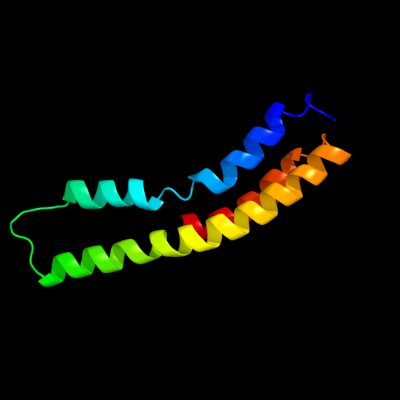
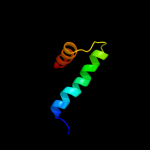
PDB 2bl2 chain F
Region: 98 - 194
Aligned: 97
Modelled: 97
Confidence: 34.6%
Identity: 9%
PDB header:hydrolase
Chain: F: PDB Molecule:v-type sodium atp synthase subunit k;
PDBTitle: the membrane rotor of the v-type atpase from enterococcus2 hirae
Phyre2
| 2 |
|
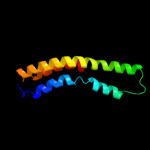
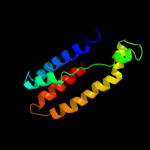
PDB 2vv5 chain D
Region: 1 - 87
Aligned: 87
Modelled: 87
Confidence: 28.3%
Identity: 14%
PDB header:membrane protein
Chain: D: PDB Molecule:small-conductance mechanosensitive channel;
PDBTitle: the open structure of mscs
Phyre2
| 3 |
|
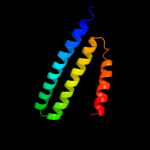
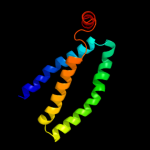
PDB 1oy8 chain A
Region: 41 - 86
Aligned: 46
Modelled: 46
Confidence: 20.3%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:acriflavine resistance protein b;
PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
Phyre2
| 4 |
|
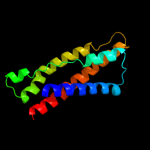
PDB 3aqp chain B
Region: 41 - 166
Aligned: 126
Modelled: 126
Confidence: 10.8%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 5 |
|
PDB 2nn6 chain H domain 3
Region: 29 - 70
Aligned: 42
Modelled: 42
Confidence: 8.1%
Identity: 17%
Fold: Eukaryotic type KH-domain (KH-domain type I)
Superfamily: Eukaryotic type KH-domain (KH-domain type I)
Family: Eukaryotic type KH-domain (KH-domain type I)
Phyre2
| 6 |
|
PDB 1iwg chain A domain 7
Region: 41 - 152
Aligned: 112
Modelled: 112
Confidence: 7.5%
Identity: 13%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 7 |
|
PDB 1pv7 chain A
Region: 4 - 106
Aligned: 103
Modelled: 103
Confidence: 7.4%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2