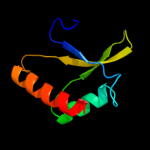
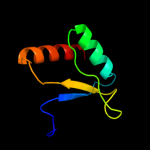
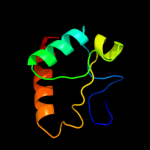
1 c1x60A_
99.7
19
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
2 d1utaa_
99.6
21
Fold: Ferredoxin-likeSuperfamily: Sporulation related repeatFamily: Sporulation related repeat3 d1wyua1
78.2
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Glycine dehydrogenase subunits (GDC-P)4 d1dq3a4
77.6
14
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Intein endonuclease5 d1m32a_
75.5
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like6 d1js3a_
71.7
8
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Pyridoxal-dependent decarboxylase7 d1iuga_
71.0
12
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like8 c3k40B_
69.1
11
PDB header: lyaseChain: B: PDB Molecule: aromatic-l-amino-acid decarboxylase;PDBTitle: crystal structure of drosophila 3,4-dihydroxyphenylalanine2 decarboxylase
9 c2yrrA_
68.8
17
PDB header: transferaseChain: A: PDB Molecule: aminotransferase, class v;PDBTitle: hypothetical alanine aminotransferase (tth0173) from thermus2 thermophilus hb8
10 d1pmma_
68.4
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Pyridoxal-dependent decarboxylase11 c3caiA_
67.8
13
PDB header: transferaseChain: A: PDB Molecule: possible aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis rv3778c2 protein
12 c3f0hA_
65.2
12
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase (rer070207000802) from2 eubacterium rectale at 1.70 a resolution
13 c3ffrA_
65.1
21
PDB header: transferaseChain: A: PDB Molecule: phosphoserine aminotransferase serc;PDBTitle: crystal structure of a phosphoserine aminotransferase serc (chu_0995)2 from cytophaga hutchinsonii atcc 33406 at 1.75 a resolution
14 d1bs0a_
62.6
20
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like15 c2fyfB_
60.0
13
PDB header: transferaseChain: B: PDB Molecule: phosphoserine aminotransferase;PDBTitle: structure of a putative phosphoserine aminotransferase from2 mycobacterium tuberculosis
16 c2zxeG_
59.1
48
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
17 c2qmaB_
57.5
8
PDB header: transferaseChain: B: PDB Molecule: diaminobutyrate-pyruvate transaminase and l-2,4-PDBTitle: crystal structure of glutamate decarboxylase domain of2 diaminobutyrate-pyruvate transaminase and l-2,4-diaminobutyrate3 decarboxylase from vibrio parahaemolyticus
18 d1m6sa_
56.5
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like19 c2dr1A_
52.0
14
PDB header: transferaseChain: A: PDB Molecule: 386aa long hypothetical serine aminotransferase;PDBTitle: crystal structure of the ph1308 protein from pyrococcus horikoshii ot3
20 c3cq6E_
51.5
11
PDB header: transferaseChain: E: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: histidinol-phosphate aminotransferase from corynebacterium2 glutamicum holo-form (plp covalently bound )
21 d1fg7a_
not modelled
49.5
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like22 c2jp3A_
not modelled
49.5
26
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
23 d2ivda2
not modelled
48.3
17
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase24 c2kncB_
not modelled
48.2
32
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
25 c3hdoB_
not modelled
46.8
20
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
26 d1qz9a_
not modelled
46.4
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like27 c3hbxB_
not modelled
46.1
21
PDB header: lyaseChain: B: PDB Molecule: glutamate decarboxylase 1;PDBTitle: crystal structure of gad1 from arabidopsis thaliana
28 d1elua_
not modelled
45.0
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like29 c2rmzA_
not modelled
43.2
32
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
30 d1v72a1
not modelled
43.1
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like31 d1t3ia_
not modelled
42.8
9
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like32 d1jf9a_
not modelled
41.4
11
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like33 c2jo1A_
not modelled
40.3
13
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
34 d1seza2
not modelled
39.8
26
Fold: FAD-linked reductases, C-terminal domainSuperfamily: FAD-linked reductases, C-terminal domainFamily: L-aminoacid/polyamine oxidase35 d1oj4a2
not modelled
39.2
14
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase IspE36 c3ly1C_
not modelled
38.8
14
PDB header: transferaseChain: C: PDB Molecule: putative histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (yp_050345.1) from erwinia carotovora atroseptica scri1043 at 1.80 a3 resolution
37 d2erra1
not modelled
38.4
7
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD38 c3getA_
not modelled
37.9
14
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (np_281508.1) from campylobacter jejuni at 2.01 a resolution
39 c3f9tB_
not modelled
36.9
29
PDB header: lyaseChain: B: PDB Molecule: l-tyrosine decarboxylase mfna;PDBTitle: crystal structure of l-tyrosine decarboxylase mfna (ec 4.1.1.25)2 (np_247014.1) from methanococcus jannaschii at 2.11 a resolution
40 d1ztpa1
not modelled
35.7
8
Fold: eIF4e-likeSuperfamily: eIF4e-likeFamily: BLES03-like41 c3a2bA_
not modelled
35.6
14
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
42 c2okkA_
not modelled
35.2
13
PDB header: lyaseChain: A: PDB Molecule: glutamate decarboxylase 2;PDBTitle: the x-ray crystal structure of the 65kda isoform of glutamic acid2 decarboxylase (gad65)
43 d2bwna1
not modelled
34.9
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like44 c2huuA_
not modelled
33.6
15
PDB header: transferaseChain: A: PDB Molecule: alanine glyoxylate aminotransferase;PDBTitle: crystal structure of aedes aegypti alanine glyoxylate2 aminotransferase in complex with alanine
45 d1eg5a_
not modelled
33.5
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like46 d1nxia_
not modelled
33.2
13
Fold: Ferredoxin-likeSuperfamily: Hypothetical protein VC0424Family: Hypothetical protein VC042447 c2o01D_
not modelled
32.7
13
PDB header: photosynthesisChain: D: PDB Molecule: photosystem i reaction center subunit ii,PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
48 c3hqtB_
not modelled
32.4
12
PDB header: transferaseChain: B: PDB Molecule: cai-1 autoinducer synthase;PDBTitle: plp-dependent acyl-coa transferase cqsa
49 d3erja1
not modelled
30.6
26
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II50 d1x4ea1
not modelled
30.6
20
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD51 d2gova1
not modelled
30.1
7
Fold: Probable bacterial effector-binding domainSuperfamily: Probable bacterial effector-binding domainFamily: SOUL heme-binding protein52 c2w8wA_
not modelled
30.1
15
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: n100y spt with plp-ser
53 c2k8eA_
not modelled
29.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein yegp;PDBTitle: solution nmr structure of protein of unknown function yegp from e.2 coli. ontario center for structural proteomics target ec0640_1_1233 northeast structural genomics consortium target et102.
54 c2jisA_
not modelled
28.9
15
PDB header: lyaseChain: A: PDB Molecule: cysteine sulfinic acid decarboxylase;PDBTitle: human cysteine sulfinic acid decarboxylase (csad) in2 complex with plp.
55 c2dh9A_
not modelled
27.6
12
PDB header: rna binding proteinChain: A: PDB Molecule: heterogeneous nuclear ribonucleoprotein m;PDBTitle: solution structure of the c-terminal rna binding domain in2 heterogeneous nuclear ribonucleoprotein m
56 d2f8ja1
not modelled
26.4
21
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like57 d1h0ca_
not modelled
26.1
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like58 c1xtyD_
not modelled
25.8
19
PDB header: hydrolaseChain: D: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna2 hydrolase
59 c3r8kB_
not modelled
25.5
12
PDB header: apoptosisChain: B: PDB Molecule: heme-binding protein 2;PDBTitle: crystal structure of human soul protein (hexagonal form)
60 d1rk8a_
not modelled
25.3
9
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD61 d2cpia1
not modelled
23.7
4
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD62 c2hzpA_
not modelled
23.1
11
PDB header: hydrolaseChain: A: PDB Molecule: kynureninase;PDBTitle: crystal structure of homo sapiens kynureninase
63 c3p3dA_
not modelled
22.8
19
PDB header: nuclear proteinChain: A: PDB Molecule: nucleoporin 53;PDBTitle: crystal structure of the nup53 rrm domain from pichia guilliermondii
64 c3bpqD_
not modelled
22.4
31
PDB header: toxinChain: D: PDB Molecule: toxin rele3;PDBTitle: crystal structure of relb-rele antitoxin-toxin complex from2 methanococcus jannaschii
65 c2zv3E_
not modelled
22.1
23
PDB header: hydrolaseChain: E: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
66 c2d3kA_
not modelled
21.9
17
PDB header: hydrolaseChain: A: PDB Molecule: peptidyl-trna hydrolase;PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
67 c2eqkA_
not modelled
21.9
10
PDB header: transcriptionChain: A: PDB Molecule: tudor domain-containing protein 4;PDBTitle: solution structure of the tudor domain of tudor domain-2 containing protein 4
68 d1mdoa_
not modelled
21.9
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like69 c3eucB_
not modelled
21.8
8
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase 2;PDBTitle: crystal structure of histidinol-phosphate aminotransferase2 (yp_297314.1) from ralstonia eutropha jmp134 at 2.05 a resolution
70 d2c0ra1
not modelled
20.2
7
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like71 c2z0rA_
not modelled
19.7
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha0547;PDBTitle: crystal structure of hypothetical protein ttha0547
72 d1rlka_
not modelled
19.4
28
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II73 c2q5cA_
not modelled
18.7
17
PDB header: transcriptionChain: A: PDB Molecule: ntrc family transcriptional regulator;PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
74 c2k49A_
not modelled
18.7
4
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein so_3888;PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
75 c3ns5B_
not modelled
18.6
18
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 3 subunit b;PDBTitle: crystal structure of the rna recognition motif of yeast eif3b residues2 76-161
76 c3hdeA_
not modelled
18.2
14
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme;PDBTitle: crystal structure of full-length endolysin r21 from phage 21
77 d1wi6a1
not modelled
18.1
12
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD78 c1no8A_
not modelled
17.8
11
PDB header: rna binding proteinChain: A: PDB Molecule: aly;PDBTitle: solution structure of the nuclear factor aly rbd domain
79 d1no8a_
not modelled
17.8
11
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD80 c2wseD_
not modelled
17.4
13
PDB header: photosynthesisChain: D: PDB Molecule: photosystem i reaction center subunit ii,PDBTitle: improved model of plant photosystem i
81 c2jwaA_
not modelled
17.4
25
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
82 c2pbiA_
not modelled
17.3
9
PDB header: signaling proteinChain: A: PDB Molecule: regulator of g-protein signaling 9;PDBTitle: the multifunctional nature of gbeta5/rgs9 revealed from its crystal2 structure
83 c3ml6D_
not modelled
16.8
12
PDB header: protein transportChain: D: PDB Molecule: chimeric complex between protein dishevlled2 homolog dvl-2PDBTitle: a complex between dishevlled2 and clathrin adaptor ap-2
84 d1gcca_
not modelled
16.2
6
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: GCC-box binding domain85 c2divA_
not modelled
16.0
10
PDB header: rna binding proteinChain: A: PDB Molecule: trna selenocysteine associated protein;PDBTitle: solution structure of the rrm domain of trna selenocysteine2 associated protein
86 c3ht4B_
not modelled
16.0
15
PDB header: lyaseChain: B: PDB Molecule: aluminum resistance protein;PDBTitle: crystal structure of the q81a77_baccr protein from bacillus2 cereus. northeast structural genomics consortium target3 bcr213
87 d1wg1a_
not modelled
15.9
8
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD88 d1q7sa_
not modelled
15.7
19
Fold: Peptidyl-tRNA hydrolase IISuperfamily: Peptidyl-tRNA hydrolase IIFamily: Peptidyl-tRNA hydrolase II89 c3ffhA_
not modelled
15.6
20
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: the crystal structure of histidinol-phosphate aminotransferase from2 listeria innocua clip11262.
90 d1jb0d_
not modelled
15.0
10
Fold: Photosystem I subunit PsaDSuperfamily: Photosystem I subunit PsaDFamily: Photosystem I subunit PsaD91 c2ordA_
not modelled
15.0
13
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
92 c3n23E_
not modelled
14.6
24
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
93 d2cqha1
not modelled
14.6
15
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD94 d2ch1a1
not modelled
14.4
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like95 d2cpea1
not modelled
13.8
8
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD96 c2l9vA_
not modelled
13.8
14
PDB header: rna binding proteinChain: A: PDB Molecule: pre-mrna-processing factor 40 homolog a;PDBTitle: nmr structure of the ff domain l24a mutant's folding transition state
97 d2cpha1
not modelled
13.8
19
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD98 c3e9kA_
not modelled
13.7
11
PDB header: hydrolaseChain: A: PDB Molecule: kynureninase;PDBTitle: crystal structure of homo sapiens kynureninase-3-hydroxyhippuric acid2 inhibitor complex
99 d1fxla1
not modelled
13.6
9
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD